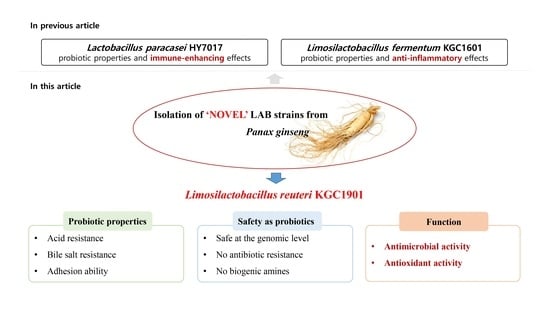
Characterization of Limosilactobacillus reuteri KGC1901 Newly Isolated from Panax ginseng Root as a Probiotic and Its Safety Assessment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of LAB Strains from Raw Ginseng and Identification of L. reuteri KGC1901
2.2. Carbohydrate Utilization and Enzymatic Activities
2.3. Acid and Bile Salt Resistance
2.4. Intestinal Adhesion and Mucin Binding Ability
2.5. Immunofluorescence Staining
2.6. Antibiotic Resistance and Virulence Factor
2.7. Determination of Hemolytic Activity and Biogenic Amine
2.8. Antimicrobial Activity of L. reuteri KGC1901 CFS against Clostridium difficile
2.9. DPPH Radical Scavenging Activity
2.10. Cell Culture and Nitric Oxide (NO) Production Assay
3. Results and Discussion
3.1. Isolation and Identification of L. reuteri KGC1901 from Ginseng Root
3.2. Carbohydrate Fermentation and Enzymatic Activities
3.3. Probiotic Properties
3.4. Safety Assessments
3.5. Antimicrobial Activity against Clostridium difficile
3.6. Antioxidant Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The human microbiome and its impacts on health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of action of probiotics. Adv. Nutr. 2019, 10, S49–S66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, H.; Beresford, T.P.; Cotter, P.D. Health benefits of lactic acid bacteria (LAB) fermentates. Nutrients 2020, 12, 1679. [Google Scholar] [CrossRef]
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Lee, Y.-S.; Yu, H.-Y.; Kwon, M.; Kim, K.-K.; In, G.; Hong, S.-K.; Kim, S.-K. Anti-inflammatory effects of Limosilactobacillus fermentum KGC1601 isolated from panax ginseng and its probiotic characteristics. Foods 2022, 11, 1707. [Google Scholar] [CrossRef]
- Li, H.-Y.; Zhou, D.-D.; Gan, R.-Y.; Huang, S.-Y.; Zhao, C.-N.; Shang, A.; Xu, X.-Y.; Li, H.-B. Effects and mechanisms of probiotics, prebiotics, synbiotics, and postbiotics on metabolic diseases targeting gut microbiota: A narrative review. Nutrients 2021, 13, 3211. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics regulate gut microbiota: An effective method to improve immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- Lee, H.; Jung, K.B.; Kwon, O.; Son, Y.S.; Choi, E.; Yu, W.D.; Son, N.; Jeon, J.H.; Jo, H.; Yang, H. Limosilactobacillus reuteri DS0384 promotes intestinal epithelial maturation via the postbiotic effect in human intestinal organoids and infant mice. Gut Microbes 2022, 14, 2121580. [Google Scholar] [CrossRef]
- Doron, S.; Snydman, D.R. Risk and safety of probiotics. Clin. Infect. Dis. 2015, 60, S129–S134. [Google Scholar] [CrossRef] [Green Version]
- Hotel, A.C.P.; Cordoba, A. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Prevention 2001, 5, 1–10. [Google Scholar]
- Vitali, B.; Minervini, G.; Rizzello, C.G.; Spisni, E.; Maccaferri, S.; Brigidi, P.; Gobbetti, M.; Di Cagno, R. Novel probiotic candidates for humans isolated from raw fruits and vegetables. Food Microbiol. 2012, 31, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Li, K.; Xiao, Y.; Zhong, A.; Tang, J.; Duan, Y.; Li, Z. Limosilactobacillus reuteri Regulating Intestinal Function: A Review. Fermentation 2023, 9, 19. [Google Scholar] [CrossRef]
- Arshad, F.; Mehmood, R.; Hussain, S.; Khan, M.A.; Khan, M. Lactobacilli as probiotics and their isolation from different sources. Br. J. Res. 2018, 5, 43. [Google Scholar] [CrossRef]
- Hyun, S.H.; Bhilare, K.D.; In, G.; Park, C.-K.; Kim, J.-H. Effects of Panax ginseng and ginsenosides on oxidative stress and cardiovascular diseases: Pharmacological and therapeutic roles. J. Ginseng Res. 2022, 46, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.L.; Bae, H. Bacterial endophytes from ginseng and their biotechnological application. J. Ginseng Res. 2022, 46, 1–10. [Google Scholar] [CrossRef]
- Hua, M.; Fan, M.; Li, Z.; Sha, J.; Li, S.; Sun, Y. Ginseng soluble dietary fiber can regulate the intestinal flora structure, promote colon health, affect appetite and glucolipid metabolism in rats. J. Funct. Foods 2021, 83, 104534. [Google Scholar] [CrossRef]
- Yu, H.-Y.; Rhim, D.-B.; Kim, S.-K.; Ban, O.-H.; Oh, S.-K.; Seo, J.; Hong, S.-K. Growth promotion effect of red ginseng dietary fiber to probiotics and transcriptome analysis of Lactiplantibacillus plantarum. J. Ginseng Res. 2023, 47, 159–165. [Google Scholar] [CrossRef]
- Mo, S.-J.; Nam, B.; Bae, C.-H.; Park, S.-D.; Shim, J.-J.; Lee, J.-L. Characterization of novel Lactobacillus paracasei HY7017 capable of improving physiological properties and immune enhancing effects using red ginseng extract. Fermentation 2021, 7, 238. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Yu, H.-Y.; Kwon, M.; Lee, S.-H.; Park, J.-I.; Seo, J.; Kim, S.-K. Probiotic Characteristics and Safety Assessment of Lacticaseibacillus casei KGC1201 Isolated from Panax ginseng. J. Microbiol. Biotechnol. 2023, 33, 1–8. [Google Scholar] [CrossRef]
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile infection. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, e1–e48. [Google Scholar] [CrossRef] [PubMed]
- Tartof, S.Y.; Rieg, G.K.; Wei, R.; Tseng, H.F.; Jacobsen, S.J.; Kalvin, C.Y. A comprehensive assessment across the healthcare continuum: Risk of hospital-associated Clostridium difficile infection due to outpatient and inpatient antibiotic exposure. Infect. Control Hosp. Epidemiol. 2015, 36, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Kalakuntla, A.S.; Nalakonda, G.; Nalakonda, K.; Pidikiti, C.V.; Aasim, S.A. Probiotics and Clostridium difficile: A review of dysbiosis and the rehabilitation of gut microbiota. Cureus 2019, 11, e5063. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.F.; Tseng, K.C.; Chiang, S.S.; Lee, B.H.; Hsu, W.H.; Pan, T.M. Immunomodulatory and antioxidant potential of Lactobacillus exopolysaccharides. J. Sci. Food Agric. 2011, 91, 2284–2291. [Google Scholar] [CrossRef]
- Tang, W.; Li, C.; He, Z.; Pan, F.; Pan, S.; Wang, Y. Probiotic properties and cellular antioxidant activity of Lactobacillus plantarum MA2 isolated from Tibetan kefir grains. Probiotics Antimicrob. Proteins 2018, 10, 523–533. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.-S.; Kim, Y.; Jeong, Y.; Kim, J.-E.; Paek, N.-S.; Kang, C.-H. Antioxidant and probiotic properties of Lactobacilli and Bifidobacteria of human origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- Fessard, A.; Bourdon, E.; Payet, B.; Remize, F. Identification, stress tolerance, and antioxidant activity of lactic acid bacteria isolated from tropically grown fruits and leaves. Can. J. Microbiol. 2016, 62, 550–561. [Google Scholar] [CrossRef]
- Averina, O.V.; Poluektova, E.U.; Marsova, M.V.; Danilenko, V.N. Biomarkers and utility of the antioxidant potential of probiotic Lactobacilli and Bifidobacteria as representatives of the human gut microbiota. Biomedicines 2021, 9, 1340. [Google Scholar] [CrossRef]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T. In vitro probiotic potential and safety evaluation (hemolytic, cytotoxic activity) of Bifidobacterium strains isolated from raw camel milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.F.; Brown, L. Evaluation of the E test, a novel method of quantifying antimicrobial activity. J. Antimicrob. Chemother. 1991, 27, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder–an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Bang, W.Y.; Ban, O.-H.; Lee, B.S.; Oh, S.; Park, C.; Park, M.-K.; Jung, S.K.; Yang, J.; Jung, Y.H. Genomic-, phenotypic-, and toxicity-based safety assessment and probiotic potency of Bacillus coagulans IDCC 1201 isolated from green malt. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab026. [Google Scholar] [CrossRef]
- Lyons, C.R.; Orloff, G.; Cunningham, J. Molecular cloning and functional expression of an inducible nitric oxide synthase from a murine macrophage cell line. J. Biol. Chem. 1992, 267, 6370–6374. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Cole, C.; Fuller, R. The effect of dietary fat and yoghurt on colonic bacterial enzymes (β-glucosidase and β-glucuronidase) associated with colon cancer. Food Microbiol. 1987, 4, 77–81. [Google Scholar] [CrossRef]
- Kim, D.-H.; Jin, Y.-H. Intestinal bacterial β-glucuronidase activity of patients with colon cancer. Arch. Pharm. Res. 2001, 24, 564–567. [Google Scholar] [CrossRef]
- Ayyash, M.M.; Abdalla, A.K.; AlKalbani, N.S.; Baig, M.A.; Turner, M.S.; Liu, S.-Q.; Shah, N.P. Invited review: Characterization of new probiotics from dairy and nondairy products—Insights into acid tolerance, bile metabolism and tolerance, and adhesion capability. J. Dairy Sci. 2021, 104, 8363–8379. [Google Scholar] [CrossRef]
- Castro-López, C.; Romero-Luna, H.E.; García, H.S.; Vallejo-Cordoba, B.; González-Córdova, A.F.; Hernández-Mendoza, A. Key Stress Response Mechanisms of Probiotics during Their Journey Through the Digestive System: A Review. Probiotics Antimicrob. Proteins 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Manini, F.; Casiraghi, M.; Poutanen, K.; Brasca, M.; Erba, D.; Plumed-Ferrer, C. Characterization of lactic acid bacteria isolated from wheat bran sourdough. LWT-Food Sci. Technol. 2016, 66, 275–283. [Google Scholar] [CrossRef]
- Fu, X.; Lyu, L.; Wang, Y.; Zhang, Y.; Guo, X.; Chen, Q.; Liu, C. Safety assessment and probiotic characteristics of Enterococcus lactis JDM1. Microb. Pathog. 2022, 163, 105380. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.; Lee, S.Y. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64, S95–S100. [Google Scholar] [CrossRef] [Green Version]
- Sudo, N. Biogenic amines: Signals between commensal microbiota and gut physiology. Front. Endocrinol. 2019, 10, 504. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Capillas, C.; Herrero, A.M. Impact of biogenic amines on food quality and safety. Foods 2019, 8, 62. [Google Scholar] [CrossRef] [Green Version]
- Gardini, F.; Özogul, Y.; Suzzi, G.; Tabanelli, G.; Özogul, F. Technological factors affecting biogenic amine content in foods: A review. Front. Microbiol. 2016, 7, 1218. [Google Scholar] [CrossRef] [Green Version]
- Wójcik, W.; Łukasiewicz, M.; Puppel, K. Biogenic amines: Formation, action and toxicity—A review. J. Sci. Food Agric. 2021, 101, 2634–2640. [Google Scholar] [CrossRef]
- Pal, R.; Athamneh, A.I.; Deshpande, R.; Ramirez, J.A.; Adu, K.T.; Muthuirulan, P.; Pawar, S.; Biazzo, M.; Apidianakis, Y.; Sundekilde, U.K. Probiotics: Insights and new opportunities for Clostridioides difficile intervention. Crit. Rev. Microbiol. 2022, in press. [Google Scholar] [CrossRef]
- Engevik, M.A.; Danhof, H.A.; Shrestha, R.; Chang-Graham, A.L.; Hyser, J.M.; Haag, A.M.; Mohammad, M.A.; Britton, R.A.; Versalovic, J.; Sorg, J.A. Reuterin disrupts Clostridioides difficile metabolism and pathogenicity through reactive oxygen species generation. Gut Microbes 2020, 12, 1795388. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Auchtung, T.A.; Hermans, K.E.; Whitehead, D.; Borhan, B.; Britton, R.A. The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 2010, 156, 1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann-Schaal, M.; Metzendorf, N.G.; Troitzsch, D.; Nuss, A.M.; Hofmann, J.D.; Beckstette, M.; Dersch, P.; Otto, A.; Sievers, S. Tracking gene expression and oxidative damage of O2-stressed Clostridioides difficile by a multi-omics approach. Anaerobe 2018, 53, 94–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.-C.; Hu, Z.-Y.; Li, D.-D.; Chen, Y.-X.; Xi, J.-H.; Zhao, C.-H. Application of the Reuterin System as Food Preservative or Health-Promoting Agent: A Critical Review. Foods 2022, 11, 4000. [Google Scholar] [CrossRef]
- Spinler, J.K.; Auchtung, J.; Brown, A.; Boonma, P.; Oezguen, N.; Ross, C.L.; Luna, R.A.; Runge, J.; Versalovic, J.; Peniche, A. Next-generation probiotics targeting Clostridium difficile through precursor-directed antimicrobial biosynthesis. Infect. Immun. 2017, 85, e00303–e00317. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Gutierrez-Maddox, N.; Mutukumira, A.N.; Maddox, I.S.; Shu, Q. Influence of Operating Conditions on Reuterin Production Using Resting Cells of Limosilactobacillus reuteri DPC16. Fermentation 2022, 8, 227. [Google Scholar] [CrossRef]
- Morita, H.; Toh, H.; Fukuda, S.; Horikawa, H.; Oshima, K.; Suzuki, T.; Murakami, M.; Hisamatsu, S.; Kato, Y.; Takizawa, T. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 2008, 15, 151–161. [Google Scholar] [CrossRef]
- Greppi, A.; Asare, P.T.; Schwab, C.; Zemp, N.; Stephan, R.; Lacroix, C. Isolation and comparative genomic analysis of reuterin-producing Lactobacillus reuteri from the chicken gastrointestinal tract. Front. Microbiol. 2020, 11, 1166. [Google Scholar] [CrossRef]
- Yang, K.M.; Kim, J.-S.; Kim, H.-S.; Kim, Y.-Y.; Oh, J.-K.; Jung, H.-W.; Park, D.-S.; Bae, K.-H. Lactobacillus reuteri AN417 cell-free culture supernatant as a novel antibacterial agent targeting oral pathogenic bacteria. Sci. Rep. 2021, 11, 1631. [Google Scholar] [CrossRef]







| Items | Results |
|---|---|
| Genome size (bp) | 1,963,544 |
| GC contents (%) | 38.7 |
| CDS | 1944 |
| No. of rRNA genes | 10 |
| No. of tRNA genes | 61 |
| Carbohydrates | Strains | Carbohydrates | Strains | ||
|---|---|---|---|---|---|
| KGC 1901 | KCTC 3594 | KGC 1901 | KCTC 3594 | ||
| Glycerol | - * | - | Salicin | - | - |
| Erythritol | - | - | D-cellobiose | - | - |
| D-arabinose | - | - | D-maltose | + | + |
| L-arabinose | + | + | D-lactose | + | + |
| D-ribose | + | + | D-melibiose | + | + |
| D-xylose | - | - | D-saccharose(sucrose) | + | + |
| L-xylose | - | - | D-trehalose | - | - |
| D-adonitol | - | - | Inulin | - | - |
| Methyl-βD-Xylopyranoside | - | - | D-melezitose | - | - |
| D-galactose | + | + | D-raffinose | + | + |
| D-glucose | + | + | Amidon (starch) | - | - |
| D-fructose | - | - | Glycogen | - | - |
| D-mannose | + | - | Xylitol | - | - |
| L-sorbose | - | - | Gentiobiose | - | - |
| L-rhamnose | - | - | D-turanose | - | - |
| Dulcitol | - | - | D-lyxose | - | - |
| Inositol | - | - | D-tagatose | - | - |
| D-mannitol | - | - | D-fucose | - | - |
| D-sorbitol | - | - | L-fucose | - | - |
| MethylαD-mannopyranoside | - | - | D-arabitol | - | - |
| MethylαD-Glucopyranoside | - | - | L-arabitol | - | - |
| N-acetylglucosamine | - | - | Potassium gluconate | v | v |
| Amygdalin | - | - | Potassium 2-ketogluconate | - | - |
| Arbutin | - | - | Potassium 5-ketogluconate | - | - |
| Esculin ferric citrate | + | + | |||
| Enzymes | KGC1901 | Enzymes | KGC1901 |
|---|---|---|---|
| Alkaline phosphatase | - * | Naphthol-AS-BI-phosphohydrolase | + |
| Esterase | + | α-Galactosidase | + |
| Esterase lipase | - | β-Galactosidase | +++ |
| Lipase | - | β-Glucuronidase | - |
| Leucine arylamidase | +++ | α-Glucosidase | + |
| Valine arylamidase | + | β-Glucosidase | - |
| Crystinearylamidase | - | N-acetyl-β-glucosaminidase | - |
| Trypsin | - | α-Mannosidase | - |
| α-Chymotrypsin | - | α-Fucosidase | - |
| Acid phosphatase | + |
| Characteristics | Survival Rate (%) | ||
|---|---|---|---|
| KGC1901 | LGG | ||
| Acid resistance | 0 h (log CFU/mL) | 8.26 ± 0.00 | 8.03 ± 0.06 |
| 3 h (log CFU/mL) | 6.78 ± 0.11 | 6.77 ± 0.13 | |
| Survival rate (%) | 81.97 ± 1.38 | 84.19 ± 2.33 | |
| Bile salt tolerance | 0 h (log CFU/mL) | 8.26 ± 0.00 | 8.03 ± 0.06 |
| 3 h (log CFU/mL) | 7.61 ± 0.10 | 7.07 ± 0.02 | |
| Survival rate (%) | 92.04 ± 1.19 | 88.05 ± 0.96 | |
| DPPH radical scavenging (%) | 13 | 9 | |
| Antibiotic 1 | AMP | GEN | KAN | STR | ERY | CLI | TET | CHL |
|---|---|---|---|---|---|---|---|---|
| Cut-off value (mg/L) 2 | 2 | 8 | 64 | 64 | 1 | 1 | 16 | 4 |
| Observed MICs | 0.016 | 3 | 48 | 8 | 0.125 | <0.016 | 1.0–1.5 | 0.75 |
| Assessment | S 3 | S | S | S | S | S | S | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.-Y.; Kwon, M.; Lee, Y.-S.; Lee, S.-H.; Kim, S.-K. Characterization of Limosilactobacillus reuteri KGC1901 Newly Isolated from Panax ginseng Root as a Probiotic and Its Safety Assessment. Fermentation 2023, 9, 228. https://doi.org/10.3390/fermentation9030228
Yu H-Y, Kwon M, Lee Y-S, Lee S-H, Kim S-K. Characterization of Limosilactobacillus reuteri KGC1901 Newly Isolated from Panax ginseng Root as a Probiotic and Its Safety Assessment. Fermentation. 2023; 9(3):228. https://doi.org/10.3390/fermentation9030228
Chicago/Turabian StyleYu, Hye-Young, Mijin Kwon, Yun-Seok Lee, Seung-Ho Lee, and Sang-Kyu Kim. 2023. "Characterization of Limosilactobacillus reuteri KGC1901 Newly Isolated from Panax ginseng Root as a Probiotic and Its Safety Assessment" Fermentation 9, no. 3: 228. https://doi.org/10.3390/fermentation9030228
APA StyleYu, H.-Y., Kwon, M., Lee, Y.-S., Lee, S.-H., & Kim, S.-K. (2023). Characterization of Limosilactobacillus reuteri KGC1901 Newly Isolated from Panax ginseng Root as a Probiotic and Its Safety Assessment. Fermentation, 9(3), 228. https://doi.org/10.3390/fermentation9030228