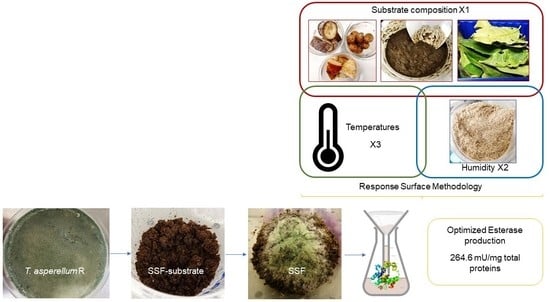
Optimization of Esterase Production in Solid-State Fermentation of Agricultural Digestate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Species
2.2. Organic Wastes
2.3. Production of Enzymes in Solid-State Fermentation
2.3.1. Substrate Composition
2.3.2. Crude Extract Collection
2.3.3. Cellulase Activity
2.3.4. Esterase Activity
2.4. Optimization of Esterase Production in Solid-State Fermentation
2.5. Response Surface Methodology
3. Results and Discussion
3.1. Cellulase and Esterase Activity in Solid-State Fermentation
3.2. Light Does Not Influence Esterase Activity
3.3. Esterase Production Optimization
3.3.1. Fungal Growth
3.3.2. The Model Fitness
- Digestate 70%:
- Digestate 50%:
3.3.3. Esterase Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bornscheuer, U.T. Microbial Carboxyl Esterases: Classification, Properties and Application in Biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Rafeeq, H.; Hussain, A.; Shabbir, S.; Ali, S.; Bilal, M.; Sher, F.; Iqbal, H.M.N. Esterases as Emerging Biocatalysts: Mechanistic Insights, Genomic and Metagenomic, Immobilization, and Biotechnological Applications. Biotechnol. Appl. Biochem. 2022, 69, 2176–2194. [Google Scholar] [CrossRef] [PubMed]
- De Bhowmick, G.; Sarmah, A.K.; Sen, R. Lignocellulosic Biorefinery as a Model for Sustainable Development of Biofuels and Value Added Products. Bioresour. Technol. 2018, 247, 1144–1154. [Google Scholar] [CrossRef]
- Contesini, F.J.; Frandsen, R.J.N.; Damasio, A. Editorial: CAZymes in Biorefinery: From Genes to Application. Front. Bioeng. Biotechnol. 2021, 9, 622817. [Google Scholar] [CrossRef]
- Calvo-Flores, F.G.; Martin-Martinez, F.J. Biorefineries: Achievements and Challenges for a Bio-Based Economy. Front. Chem. 2022, 10, 973417. [Google Scholar] [CrossRef]
- De Buck, V.; Polanska, M.; Van Impe, J. Modeling Biowaste Biorefineries: A Review. Front. Sustain. Food Syst. 2020, 4, 11. [Google Scholar] [CrossRef]
- Teigiserova, D.A.; Bourgine, J.; Thomsen, M. Closing the Loop of Cereal Waste and Residues with Sustainable Technologies: An Overview of Enzyme Production via Fungal Solid-State Fermentation. Sustain. Prod. Consum. 2021, 27, 845–857. [Google Scholar] [CrossRef]
- Pitol, L.O.; Finkler, A.T.J.; Dias, G.S.; Machado, A.S.; Zanin, G.M.; Mitchell, D.A.; Krieger, N. Optimization Studies to Develop a Low-Cost Medium for Production of the Lipases of Rhizopus Microsporus by Solid-State Fermentation and Scale-up of the Process to a Pilot Packed-Bed Bioreactor. Process Biochem. 2017, 62, 37–47. [Google Scholar] [CrossRef]
- Savino, S.; Bulgari, D.; Monti, E.; Gobbi, E. Agro-Industrial Wastes: A Substrate for Multi-Enzymes Production by Cryphonectria Parasitica. Fermentation 2021, 7, 279. [Google Scholar] [CrossRef]
- Bulgari, D.; Alias, C.; Peron, G.; Ribaudo, G.; Gianoncelli, A.; Savino, S.; Boureghda, H.; Bouznad, Z.; Monti, E.; Gobbi, E. Solid-State Fermentation of Trichoderma Spp.: A New Way to Valorize the Agricultural Digestate and Produce Value-Added Bioproducts. J. Agric. Food Chem. 2023, 71, 3994–4004. [Google Scholar] [CrossRef]
- Oliveira, F.; Abrunhosa, L.; Venâncio, A.; Belo, I.; Pérez-Rodríguez, N.; Domínguez, J.M. Aspergillus Ibericus Lipase Production by Solid-State Fermentation of Olive Pomace. In Wastes: Solutions, Treatments and Opportunities; Taylor & Francis Group: London, UK, 2015; pp. 195–201. [Google Scholar]
- Vohra, A.; Satyanarayana, T. Statistical Optimization of the Medium Components by Response Surface Methodology to Enhance Phytase Production by Pichia Anomala. Process Biochem. 2002, 37, 999–1004. [Google Scholar] [CrossRef]
- Woo, S.L.; Hermosa, R.; Lorito, M.; Monte, E. Trichoderma: A Multipurpose, Plant-Beneficial Microorganism for Eco-Sustainable Agriculture. Nat. Rev. Microbiol. 2022, 21, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Alias, C.; Bulgari, D.; Gobbi, E. It Works! Organic-Waste-Assisted Trichoderma Spp. Solid-State Fermentation on Agricultural Digestate. Microorganisms 2022, 10, 164. [Google Scholar] [CrossRef] [PubMed]
- Ghose, T.K. Measurement of Cellulase Activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Wheeler, A.B.; Braun, M.J. Package ‘ AlgDesign ’ NeedsCompilation Yes. R Proj. Stat. Comput. 2022, 1, 1–25. [Google Scholar]
- Lenth, R. V Response-Surface Methods in R, Using Rsm. J. Stat. Softw. 2009, 32, 1–17. [Google Scholar] [CrossRef]
- Kamm, B.; Kamm, M. Principles of Biorefineries. Appl. Microbiol. Biotechnol. 2004, 64, 137–145. [Google Scholar] [CrossRef]
- Schmoll, M. Assessing the Relevance of Light for Fungi: Implications and Insights into the Network of Signal Transmission. Adv. Appl. Microbiol. 2011, 76, 27–78. [Google Scholar] [CrossRef]
- Schmoll, M. Regulation of Plant Cell Wall Degradation by Light in Trichoderma. Fungal Biol. Biotechnol. 2018, 5, 10. [Google Scholar] [CrossRef]
- Ye, L.; Zhao, X.; Bao, E.; Li, J.; Zou, Z.; Cao, K. Bio-Organic Fertilizer with Reduced Rates of Chemical Fertilization Improves Soil Fertility and Enhances Tomato Yield and Quality. Sci. Rep. 2020, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Singhania, R.R.; Patel, A.K.; Soccol, C.R.; Pandey, A. Recent Advances in Solid-State Fermentation. Biochem. Eng. J. 2009, 44, 13–18. [Google Scholar] [CrossRef]
- Yafetto, L. Heliyon Application of Solid-State Fermentation by Microbial Biotechnology for Bioprocessing of Agro-Industrial Wastes from 1970 to 2020: A Review and Bibliometric Analysis. Heliyon 2022, 8, e09173. [Google Scholar] [CrossRef]
- Benjamin, S.; Pandey, A. Isolation and Characterization of Three Distinct Forms of Lipases from Candida Rugosa Produced in Solid State Fermentation. Braz. Arch. Biol. Technol. 2001, 44, 213–221. [Google Scholar] [CrossRef]
- Rodriguez, J.A.; Mateos, J.C.; Nungaray, J.; González, V.; Bhagnagar, T.; Roussos, S.; Cordova, J.; Baratti, J. Improving Lipase Production by Nutrient Source Modification Using Rhizopus Homothallicus Cultured in Solid State Fermentation. Process Biochem. 2006, 41, 2264–2269. [Google Scholar] [CrossRef]
- Shabtai, Y.; Daya-Mishne, N. Production, Purification, and Properties of a Lipase from a Bacterium (Pseudomonas Aeruginosa YS-7) Capable of Growing in Water-Restricted Environments. Appl. Environ. Microbiol. 1992, 58, 174–180. [Google Scholar] [CrossRef]
- Tawfik, A.; Eraky, M.; Alhajeri, N.S.; Osman, A.I.; Rooney, D.W. Cultivation of Microalgae on Liquid Anaerobic Digestate for Depollution, Biofuels and Cosmetics: A Review. Environ. Chem. Lett. 2022, 20, 3631–3656. [Google Scholar] [CrossRef]
- El-housseiny, G.S.; Ibrahim, A.A.; Yassien, M.A.; Aboshanab, K.M. Production and Statistical Optimization of Paromomycin by Streptomyces Rimosus NRRL 2455 in Solid State Fermentation. BMC Microbiol. 2021, 21, 34. [Google Scholar] [CrossRef]
- Singh, R.S.; Chauhan, K.; Jindal, A. Response Surface Optimization of Solid State Fermentation for Inulinase Production from Penicillium Oxalicum Using Corn. J. Food Sci. Technol. 2018, 55, 2544–2551. [Google Scholar] [CrossRef]
- Xu, W.; Wong, W.K.; Tan, K.C.; Xu, J.-X. Finding High-Dimensional D-Optimal Designs for Logistic Models via Differential Evolution. IEEE Access 2019, 7, 7133–7146. [Google Scholar] [CrossRef]
- Mahalaxmi, Y.; Sathish, T.; Subba Rao, C.; Prakasham, R.S. Corn Husk as a Novel Substrate for the Production of Rifamycin B by Isolated Amycolatopsis Sp. RSP 3 under SSF. Process Biochem. 2010, 45, 47–53. [Google Scholar] [CrossRef]
- Nema, A.; Patnala, S.H.; Mandari, V.; Kota, S.; Devarai, S.K. Production and Optimization of Lipase Using Aspergillus Niger MTCC 872 by Solid-State Fermentation. Bull. Natl. Res. Cent. 2019, 43, 82. [Google Scholar] [CrossRef]




| SSF Substrate ID | % Digestate | % Fruit | % Scraps |
|---|---|---|---|
| SSF-1 | 100 | 0 | 0 |
| SSF-2 | 70 | 30 | 0 |
| SSF-3 | 70 | 0 | 30 |
| SSF-4 | 70 | 15 | 15 |
| SSF-5 | 50 | 50 | 0 |
| SSF-6 | 50 | 0 | 50 |
| SSF-7 | 50 | 25 | 25 |
| Digestate 70% (w/w) | ||||
| ID | Variable | Level | ||
| −1 | 0 | 1 | ||
| X1 | Fruits (P. laurocerasus) | 0% (30%) | 15% (15%) | 30% (0%) |
| X2 | Substrate humidity | 10% | 20% | |
| X3 | Temperature | 26 °C | 30 °C | |
| Digestate 50% (w/w) | ||||
| ID | Variable | Level | ||
| −1 | 0 | 1 | ||
| X1 | Fruits (P. laurocerasus) | 0% (50%) | 25% (25%) | 50% (0%) |
| X2 | Substrate humidity | 10% | 20% | |
| X3 | Temperature | 26 °C | 30 °C | |
| Factor | |||
|---|---|---|---|
| N Run | X1 | X2 | X3 |
| 1 | −1 | −1 | −1 |
| 2 | −1 | −1 | 1 |
| 3 | −1 | 1 | −1 |
| 4 | −1 | 1 | 1 |
| 5 | 0 | −1 | −1 |
| 6 | 0 | −1 | 1 |
| 7 | 0 | 1 | −1 |
| 8 | 0 | 1 | 1 |
| 9 | 1 | −1 | −1 |
| 10 | 1 | −1 | 1 |
| 11 | 1 | 1 | −1 |
| 12 | 1 | 1 | 1 |
| Digestate = 70% | Digestate = 50% | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test | Df | Sum Sq | Mean Sq | F Value | Pr (>F) | Df | Sum Sq | Mean Sq | F Value | Pr (>F) |
| FO (X1, X2, X3) | 3 | 8411.8 | 2803.9 | 1.0 | 0.422 | 3 | 106,487.8 | 35,495.9 | 8.5 | 0.001 |
| TWI (X1, X2, X3) | 3 | 10,735.6 | 3578.5 | 1.3 | 0.320 | 3 | 17,986.6 | 5995.5 | 1.4 | 0.272 |
| PQ (X1) | 1 | 760.3 | 760.3 | 0.3 | 0.611 | 1 | 624.6 | 624.6 | 0.1 | 0.705 |
| Residuals | 16 | 45,255.5 | 2828.5 | 16 | 67,208.5 | 4200.5 | ||||
| Lack of fit | 4 | 23,310.6 | 5827.7 | 3.2 | 0.053 | 4 | 20,898.5 | 5224.6 | 1.4 | 0.307 |
| Pure error | 12 | 21,944.9 | 1828.7 | 12 | 46,310.0 | 3859.2 | ||||
| Digestate = 70% | Digestate = 50% | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | Estimate | Std. Error | t Value | Pr (>|t|) | Estimate | Std. Error | t Value | Pr (>|t|) |
| X1 | −8.2 | 11.1 | −0.7 | 0.472 | 59.7 | 13.5 | 4.4 | 0.000 |
| X2 | 2.4 | 10.9 | 0.2 | 0.826 | −23.2 | 13.2 | −1.8 | 0.098 |
| X3 | 16.8 | 10.9 | 1.5 | 0.142 | 22.3 | 13.2 | 1.7 | 0.112 |
| X1*X2 | −1.9 | 11.1 | −0.2 | 0.867 | 15.5 | 13.5 | 1.1 | 0.270 |
| X1*X3 | −20.6 | 11.1 | −1.9 | 0.082 | 19.2 | 13.5 | 1.4 | 0.175 |
| X2*X3 | −6.1 | 10.9 | −0.6 | 0.581 | −13.0 | 13.2 | −1.0 | 0.342 |
| X12 | 8.3 | 16.0 | 0.5 | 0.611 | 7.6 | 19.6 | 0.4 | 0.705 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulgari, D.; Renzetti, S.; Messgo-Moumene, S.; Monti, E.; Gobbi, E. Optimization of Esterase Production in Solid-State Fermentation of Agricultural Digestate. Fermentation 2023, 9, 524. https://doi.org/10.3390/fermentation9060524
Bulgari D, Renzetti S, Messgo-Moumene S, Monti E, Gobbi E. Optimization of Esterase Production in Solid-State Fermentation of Agricultural Digestate. Fermentation. 2023; 9(6):524. https://doi.org/10.3390/fermentation9060524
Chicago/Turabian StyleBulgari, Daniela, Stefano Renzetti, Saida Messgo-Moumene, Eugenio Monti, and Emanuela Gobbi. 2023. "Optimization of Esterase Production in Solid-State Fermentation of Agricultural Digestate" Fermentation 9, no. 6: 524. https://doi.org/10.3390/fermentation9060524
APA StyleBulgari, D., Renzetti, S., Messgo-Moumene, S., Monti, E., & Gobbi, E. (2023). Optimization of Esterase Production in Solid-State Fermentation of Agricultural Digestate. Fermentation, 9(6), 524. https://doi.org/10.3390/fermentation9060524