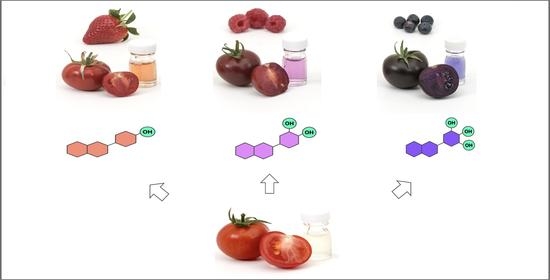
Beyond Purple Tomatoes: Combined Strategies Targeting Anthocyanins to Generate Crimson, Magenta, and Indigo Fruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Plasmid Constrzuction and Plant Transformation
2.3. Tomato Crosses and Selection of Plants
2.4. Extraction, Quantification and Metabolic Analysis of Anthocyanins
3. Results
Metabolic Engineering and Analysis of Tomatoes Producing Different Classes of Anthocyanins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Lee, Y.M.; Yoon, Y.; Yoon, H.; Park, H.M.; Song, S.; Yeum, K.J. Dietary Anthocyanins against Obesity and Inflammation. Nutrients 2017, 9, 1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, B.W.; Gong, C.C.; Song, H.F.; Cui, Y.Y. Effects of anthocyanins on the prevention and treatment of cancer. Br. J. Pharmacol. 2017, 174, 1226–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Free radicals and antioxidants: Updating a personal view. Nutr. Rev. 2012, 70, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Dangles, O.; Fenger, J.A. The Chemical Reactivity of Anthocyanins and Its Consequences in Food Science and Nutrition. Molecules 2018, 23, 1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faria, A.; Fernandes, I.; Norberto, S.; Mateus, N.; Calhau, C. Interplay between anthocyanins and gut microbiota. J. Agric. Food Chem. 2014, 62, 6898–6902. [Google Scholar] [CrossRef] [PubMed]
- Scarano, A.; Butelli, E.; De Santis, S.; Cavalcanti, E.; Hill, L.; De Angelis, M.; Giovinazzo, G.; Chieppa, M.; Martin, C.; Santino, A. Combined Dietary Anthocyanins, Flavonols, and Stilbenoids Alleviate Inflammatory Bowel Disease Symptoms in Mice. Front. Nutr. 2017, 4, 75. [Google Scholar] [CrossRef] [Green Version]
- Houghton, A.; Appelhagen, I.; Martin, C. Natural Blues: Structure Meets Function in Anthocyanins. Plants 2021, 10, 726. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Brugliera, F.; Lester, D.R.; Tanaka, Y.; Hyland, C.D.; Menting, J.G.; Lu, C.Y.; Farcy, E.; Stevenson, T.W.; Cornish, E.C. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 1993, 366, 276–279. [Google Scholar] [CrossRef]
- Halbwirth, H. The creation and physiological relevance of divergent hydroxylation patterns in the flavonoid pathway. Int. J. Mol. Sci. 2010, 11, 595–621. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Bolwell, P.G.; Bramley, P.M.; Pridham, J.B. The relative antioxidant activities of plant-derived polyphenolic flavonoids. Free Radic. Res. 1995, 22, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Bovy, A.; Bava, C.; Jaeger, S.R.; Tomes, S.; Norling, C.; Crawford, J.; Rowan, D.; McGhie, T.K.; Brendolise, C.; et al. Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol. J. 2013, 11, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Tsao, R.; Yang, R.; Young, J.C.; Zhu, H. Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC). J. Agric. Food Chem. 2003, 51, 6347–6353. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Liang, N.N.; Mu, L.; Pan, Q.H.; Wang, J.; Reeves, M.J.; Duan, C.Q. Anthocyanins and their variation in red wines I. Monomeric anthocyanins and their color expression. Molecules 2012, 17, 1571–1601. [Google Scholar] [CrossRef] [Green Version]
- Burgos, G.; Zum Felde, T.; Andre, C.; Kubow, S. The Potato and Its Contribution to the Human Diet and Health. In The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Campos, H., Ortiz, O., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 37–74. [Google Scholar]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Luo, J.; Butelli, E.; Hill, L.; Parr, A.; Niggeweg, R.; Bailey, P.; Weisshaar, B.; Martin, C. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 2008, 56, 316–326. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Alseekh, S.; Tohge, T.; Rallapalli, G.; Luo, J.; Kawar, P.G.; Hill, L.; Santino, A.; Fernie, A.R.; et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015, 6, 8635. [Google Scholar] [CrossRef] [Green Version]
- Beld, M.; Martin, C.; Huits, H.; Stuitje, A.R.; Gerats, A.G. Flavonoid synthesis in Petunia hybrida: Partial characterization of dihydroflavonol-4-reductase genes. Plant. Mol. Biol. 1989, 13, 491–502. [Google Scholar] [CrossRef]
- Fillatti, J.J.; Kiser, J.; Rose, R.; Comai, L. Efficient Transfer of a Glyphosate Tolerance Gene into Tomato Using a Binary Agrobacterium Tumefaciens Vector. Nat. Biotechnol. 1987, 5, 726–730. [Google Scholar] [CrossRef]
- Gonzali, S.; Mazzucato, A.; Perata, P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci. 2009, 14, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Mes, P.J.; Boches, P.; Myers, J.R.; Durst, R. Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hortic. Sci. 2008, 133, 262–269. [Google Scholar] [CrossRef] [Green Version]
- Tohge, T.; Zhang, Y.; Peterek, S.; Matros, A.; Rallapalli, G.; Tandron, Y.A.; Butelli, E.; Kallam, K.; Hertkorn, N.; Mock, H.P.; et al. Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J. 2015, 83, 686–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blando, F.; Berland, H.; Maiorano, G.; Durante, M.; Mazzucato, A.; Picarella, M.E.; Nicoletti, I.; Gerardi, C.; Mita, G.; Andersen, O.M. Nutraceutical Characterization of Anthocyanin-Rich Fruits Produced by “Sun Black” Tomato Line. Front. Nutr. 2019, 6, 133. [Google Scholar] [CrossRef] [Green Version]
- Bovy, A.; de Vos, R.; Kemper, M.; Schijlen, E.; Almenar Pertejo, M.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.; et al. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seitz, C.; Eder, C.; Deiml, B.; Kellner, S.; Martens, S.; Forkmann, G. Cloning, functional identification and sequence analysis of flavonoid 3’-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol. Biol. 2006, 61, 365–381. [Google Scholar] [CrossRef] [PubMed]
- De Jong, W.S.; Eannetta, N.T.; De Jong, D.M.; Bodis, M. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor. Appl. Genet. 2004, 108, 423–432. [Google Scholar] [CrossRef]
- Johnson, E.T.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Noda, N. Recent advances in the research and development of blue flowers. Breed. Sci 2018, 68, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Meyer, P.; Heidmann, I.; Forkmann, G.; Saedler, H. A New Petunia Flower Color Generated by Transformation of a Mutant with a Maize Gene. Nature 1987, 330, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Helariutta, Y.; Elomaa, P.; Kotilainen, M.; Seppanen, P.; Teeri, T.H. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol 1993, 22, 183–193. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Colanero, S.; Perata, P.; Gonzali, S. What’s behind Purple Tomatoes? Insight into the Mechanisms of Anthocyanin Synthesis in Tomato Fruits [OPEN]. Plant Physiol. 2020, 182, 1841–1853. [Google Scholar] [CrossRef] [Green Version]
- Lacombe, E.; Hawkins, S.; Van Doorsselaere, J.; Piquemal, J.; Goffner, D.; Poeydomenge, O.; Boudet, A.M.; Grima-Pettenati, J. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships. Plant J. 1997, 11, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Maloney, G.S.; DiNapoli, K.T.; Muday, G.K. The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 2014, 166, 614–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, C.; Carpenter, R.; Sommer, H.; Saedler, H.; Coen, E.S. Molecular analysis of instability in flower pigmentation of Antirrhinum majus, following isolation of the pallida locus by transposon tagging. EMBO J. 1985, 4, 1625–1630. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, V.E.; Sastry, G.R. Provocation of mutability in the level of mutation expressed at the pal-rec gene in Antirrhinum majus. Theor. Appl. Genet. 1981, 60, 303–311. [Google Scholar] [CrossRef]
- Petit, P.; Granier, T.; d’Estaintot, B.L.; Manigand, C.; Bathany, K.; Schmitter, J.M.; Lauvergeat, V.; Hamdi, S.; Gallois, B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. J. Mol. Biol. 2007, 368, 1345–1357. [Google Scholar] [CrossRef]
- Sanchez-Cabrera, M.; Jimenez-Lopez, F.J.; Narbona, E.; Arista, M.; Ortiz, P.L.; Romero-Campero, F.J.; Ramanauskas, K.; Igic, B.; Fuller, A.A.; Whittall, J.B. Changes at a Critical Branchpoint in the Anthocyanin Biosynthetic Pathway Underlie the Blue to Orange Flower Color Transition in Lysimachia arvensis. Front. Plant Sci. 2021, 12, 633979. [Google Scholar] [CrossRef]
- Adato, A.; Mandel, T.; Mintz-Oron, S.; Venger, I.; Levy, D.; Yativ, M.; Domínguez, E.; Wang, Z.; De Vos, R.C.; Jetter, R.; et al. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet. 2009, 5, e1000777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Moreno, J.P.; Tzfadia, O.; Forment, J.; Presa, S.; Rogachev, I.; Meir, S.; Orzaez, D.; Aharoni, A.; Granell, A. Characterization of a New Pink-Fruited Tomato Mutant Results in the Identification of a Null Allele of the SlMYB12 Transcription Factor. Plant Physiol. 2016, 171, 1821–1836. [Google Scholar] [CrossRef] [Green Version]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.; et al. Tomato R2R3-MYB Proteins SlANT1 and SlAN2: Same Protein Activity, Different Roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Wang, X.; Gao, J.; Guo, Y.; Huang, Z.; Du, Y. The Tomato Hoffman’s Anthocyaninless Gene Encodes a bHLH Transcription Factor Involved in Anthocyanin Biosynthesis That Is Developmentally Regulated and Induced by Low Temperatures. PLoS ONE 2016, 11, e0151067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6. [Google Scholar] [CrossRef]
- Jones, C.M.; Mes, P.; Myers, J.R. Characterization and inheritance of the Anthocyanin fruit (Aft) tomato. J. Hered. 2003, 94, 449–456. [Google Scholar] [CrossRef] [Green Version]
- Gomez Roldan, M.V.; Outchkourov, N.; van Houwelingen, A.; Lammers, M.; Romero de la Fuente, I.; Ziklo, N.; Aharoni, A.; Hall, R.D.; Beekwilder, J. An O-methyltransferase modifies accumulation of methylated anthocyanins in seedlings of tomato. Plant J. 2014, 80, 695–708. [Google Scholar] [CrossRef] [PubMed]



| ‘Crimson’ | Del/Ros1; AmDFR; f3′5′h | ||
| Peak | Rt (min) | ESI-MS (m/z) | Compound |
| 1c | 5.86 | 919 | cyanidin 3-(caffeoyl)-rutinoside-5-glucoside |
| 2c | 6.29 | 903 | pelargonidin 3-(caffeoyl)-rutinoside-5-glucoside |
| 3c | 6.57 | 933 | peonidin 3-(caffeoyl)-rutinoside-5-glucoside |
| 903 | cyanidin 3-(coumaroyl)-rutinoside-5-glucoside | ||
| 4c | 7.08 | 887 | pelargonidin 3-(coumaroyl)-rutinoside-5-glucoside |
| 5c | 7.33 | 917 | pelargonidin 3-(feruloyl)-rutinoside-5-glucoside |
| 917 | peonidin 3-(coumaroyl)-rutinoside-5-glucoside | ||
| 6c | 7.63 | 947 | peonidin 3-(feruloyl)-rutinoside-5-glucoside |
| 7c | 8.35 | 725 | pelargonidin 3-(coumaroyl)-rutinoside |
| 8c | 8.7 | 755 | pelargonidin 3-(feruloyl)-rutinoside |
| ‘Magenta’ | Del/Ros1; AmDFR; AtMYB12; f3′5′h | ||
| Peak | Rt (min) | ESI-MS (m/z) | Compound |
| 1m | 6.57 | 933 | peonidin 3-(caffeoyl)-rutinoside-5-glucoside |
| 903 | cyanidin 3-(coumaroyl)-rutinoside-5-glucoside | ||
| 2m | 7.09 | 887 | pelargonidin 3-(coumaroyl)-rutinoside-5-glucoside |
| 3m | 7.32 | 917 | peonidin 3-(coumaroyl)-rutinoside-5-glucoside |
| 4m | 7.64 | 947 | peonidin 3-(feruloyl)-rutinoside-5-glucoside |
| ‘Indigo’ | Del/Ros1; AtMYB12 | ||
| Peak | Rt (min) | ESI-MS (m/z) | Compound |
| 1i | 5.28 | 935 | delphinidin 3-(caffeoyl)-rutinoside-5-glucoside |
| 2i | 6 | 919 | delphinidin 3-(coumaroyl)-rutinoside-5-glucoside petunidin 3-(caffeoyl)-rutinoside-5-glucoside |
| 949 | |||
| 3i | 6.32 | 949 | delphinidin 3-(feruloyl)-rutinoside-5-glucoside |
| 4i | 6.71 | 933 | petunidin 3-(coumaroyl)-rutinoside-5-glucoside |
| 5i | 7.02 | 963 | petunidin 3-(feruloyl)-rutinoside-5-glucoside |
| 6i | 7.43 | 947 | malvidin 3-(coumaroyl)-rutinoside-5-glucoside |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butelli, E.; Bulling, K.; Hill, L.; Martin, C. Beyond Purple Tomatoes: Combined Strategies Targeting Anthocyanins to Generate Crimson, Magenta, and Indigo Fruit. Horticulturae 2021, 7, 327. https://doi.org/10.3390/horticulturae7090327
Butelli E, Bulling K, Hill L, Martin C. Beyond Purple Tomatoes: Combined Strategies Targeting Anthocyanins to Generate Crimson, Magenta, and Indigo Fruit. Horticulturae. 2021; 7(9):327. https://doi.org/10.3390/horticulturae7090327
Chicago/Turabian StyleButelli, Eugenio, Katharina Bulling, Lionel Hill, and Cathie Martin. 2021. "Beyond Purple Tomatoes: Combined Strategies Targeting Anthocyanins to Generate Crimson, Magenta, and Indigo Fruit" Horticulturae 7, no. 9: 327. https://doi.org/10.3390/horticulturae7090327
APA StyleButelli, E., Bulling, K., Hill, L., & Martin, C. (2021). Beyond Purple Tomatoes: Combined Strategies Targeting Anthocyanins to Generate Crimson, Magenta, and Indigo Fruit. Horticulturae, 7(9), 327. https://doi.org/10.3390/horticulturae7090327