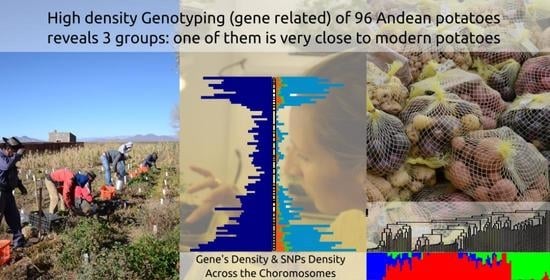
Assessment of Genetic Diversity and Relatedness in an Andean Potato Collection from Argentina by High-Density Genotyping
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Preparation
2.3. Genotyping
2.4. Diversity Analysis
2.5. Population Structure Analysis
3. Results
3.1. Genetic Diversity Analysis
3.2. Population Structure Analysis
3.3. Clustering Analysis
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. FAO Production Statistics. Food and Agriculture Organization: Crops Statistics Database. Available online: www.faostat.fao.org (accessed on 7 March 2021).
- Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.K.; Bolser, D.; de Boer, J.; Sønderkær, M.; Amoros, W.; Carboni, M.; D’Ambrosio, J.M.; de la Cruz, G.; Di Genova, A.; Douches, D.S.; et al. Construction of Reference Chromosome-Scale Pseudomolecules for Potato: Integrating the Potato Genome with Genetic and Physical Maps. G3 Genes Genomes Genet. 2013, 3, 2031–2047. [Google Scholar] [CrossRef] [Green Version]
- Spooner, D.M.; Núñez, J.; Rodríguez, F.P.; Naik, S.; Ghislain, M. Nuclear and chloroplast DNA reassessment of the origin of Indian potato varieties and its implications for the origin of the early European potato. Theor. Appl. Genet. 2005, 110, 1020–1026. [Google Scholar] [CrossRef]
- Milbourne, D.; Pande, B.; Bryan, G.J. Genome Mapping and Molecular Breeding in Plants Pulses, Sugar and Tuber Crops; Springer: Berlin/Heidelberg, Germany, 2007; Volume 12, pp. 205–236. [Google Scholar]
- Rumold, C.U.; Aldenferder, M.S. Late Archaic–Early Formative period microbotanical evidence for potato at Jiskairumoko in the Titicaca Basin of southern Peru. Proc. Natl. Acad. Sci. USA 2016, 113, 13672–13677. [Google Scholar] [CrossRef] [Green Version]
- Dodds, K.S. Classification of cultivated potatoes. In the Potato and Its Wild Relatives; Series of Botanical Studies 4; Correll, D.S., Ed.; Texas Research Foundation: Renner, TX, USA, 1962; pp. 517–539. [Google Scholar]
- Gavrilenko, T.; Antonova, O.; Shuvalova, A.; Krylova, E.; Alpatyeva, N.; Spooner, D.M.; Novikova, L. Genetic diversity and origin of cultivated potatoes based on plastid microsatellite polymorphism. Genet. Resour. Crop. Evol. 2013, 60, 1997–2015. [Google Scholar] [CrossRef]
- Vos, P.G.; Uitdewilligen, J.G.; Voorrips, R.E.; Visser, R.G.; van Eck, H.J. Development and analysis of a 20K SNP array for potato (Solanum tuberosum): An insight into the breeding history. Theor. Appl. Genet. 2015, 128, 2387–2401. [Google Scholar] [CrossRef] [Green Version]
- Ames, M.; Spooner, D.M. DNA from herbarium specimens settles a controversy about origins of the European potato. Am. J. Bot. 2008, 95, 252–257. [Google Scholar] [CrossRef] [Green Version]
- Atencio, H.M.; Ispizúa, N.V.; Feingold, S.; Clausen, A.M. Conservación ex situ de variedades de papas nativas. Caso de estudio de la variedad ‘Collareja’ del noroeste de la Argentina. Rev. Investig. Agropecu. (RIA) 2019, 45, 242–251. [Google Scholar]
- Andre, C.M.; Ghislain, M.; Bertin, P.; Oufir, M.; del Rosario Herrera, M.; Hoffmann, L.; Hausman, J.F.; Larondelle, Y.; Evers, D. Andean potato cultivars Solanum tuberosum L. as a source of antioxidant and mineral micronutrients. J. Agric. Food Chem. 2007, 55, 366–378. [Google Scholar] [CrossRef]
- Clausen, A.M. Tesoros de la Biodiversidad Andina: Las Papas Nativas y su Valor Para la Humanidad. Recuperado el 18 de Marzo de 2019, del Sitio Web del Instituto Nacional de Tecnología Agropecuaria. Available online: http://inta.gob.ar/noticias/tesoros-de-la-biodiversidad-andina-las-papas-nativas-y-su-valor-para-la-humanidad (accessed on 18 March 2021).
- Ispizúa, V.N.; Guma, I.R.; Feingold, S.; Clausen, A.M. Genetic diversity of potato landraces from northwestern Argentina assessed with simple sequence repeats (SSRs). Genet. Resour. Crop. Evol. 2007, 54, 1833–1848. [Google Scholar] [CrossRef]
- Ortiz, R. The state of the use of potato genetic diversity. In Broadening the Genetic Base of Crop Production; CABI Publishing: Wallingford, UK, 2011; pp. 181–200. [Google Scholar]
- Hardigan, M.A.; Bamberg, J.; Buell, C.R.; Douches, D.S. Taxonomy and genetic differentiation among wild and cultivated germplasm of Solanum sect. Petota. Plant Genome 2015, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Monte, M.N.; Burusco, M.F.R.; Carboni, M.F.; Castellote, M.A.; Sucar, S.; Norero, N.S.; Colman, S.L.; Massa, G.A.; Colavita, M.L.; Feingold, S.E. Genetic Diversity in Argentine Andean Potatoes by Means of Functional Markers. Am. J. Potato Res. 2018, 95, 286–300. [Google Scholar] [CrossRef]
- Colman, S.L.; Massa, G.A.; Carboni, M.F.; Feingold, S.E. Cold sweetening diversity in Andean potato germplasm from Argentina. J. Sci. Food Agric. 2019, 678, 499–524. [Google Scholar] [CrossRef] [Green Version]
- Esquinas-Alcázar, J. Protecting crop genetic diversity for food security: Political, ethical and technical challenges. Nat. Rev. Genet. 2005, 6, 946–953. [Google Scholar] [CrossRef] [PubMed]
- FAO. Tratado Internacional Sobre los Recursos Fitogenéticos Para la Alimentación y la Agricultura. 2001. Available online: http://www.fao.org/plant-treaty/es/ (accessed on 8 March 2021).
- Brush, S.B.; Taylor, J.; Bellon, M.R. Technology adoption and biological diversity in Andean potato agriculture. J. Dev. Biol. 1992, 39, 365–387. [Google Scholar] [CrossRef]
- Ortiz, R. Genomic-Led Potato Breeding for Increasing Genetic Gains: Achievements and Outlook. Crop. Breed. Genet. Genom. 2020, 2, e200010. [Google Scholar] [CrossRef]
- Bradshaw, J.E.; Hackett, C.A.; Meyer, R.C.; Milbourne, D.; McNicol, J.W.; Phillips, M.S.; Waugh, R. Identification of AFLP and SSR markers associated with quantitative resistance to Globodera pallida (Stone) in tetraploid potato (Solanum tuberosum subsp. tuberosum) with a view to marker-assisted selection. Theor. Appl. Genet. 1998, 97, 202–210. [Google Scholar]
- Ghislain, M.; Trognitz, B.; Herrera, M.D.R.; Solis, J.; Casallo, G.; Vasquez-Robinet, C.; Hurtado, O.; Castillo, R.; Portal, L.; Orrillo, M. Genetic loci associated with field resistance to late blight in offspring of Solanum phureja and S. tuberosum grown under short-day conditions. Theor. Appl. Genet. 2001, 103, 433–442. [Google Scholar] [CrossRef]
- Norero, N.; Malleville, J.; Huarte, M.; Feingold, S.E. Cost efficient potato Solanum tuberosum L. cultivar identification by microsatellite amplification. Potato Res. 2004, 45, 131–138. [Google Scholar] [CrossRef]
- Feingold, S.; Lloyd, J.; Norero, N.; Bonierbale, M.; Lorenzen, J. Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theor. Appl. Genet. 2005, 111, 456–466. [Google Scholar] [CrossRef]
- Fu, Y.B.; Peterson, G.W.; Richards, K.W.; Tarn, T.R.; Percy, J.E. Genetic diversity of Canadian and exotic potato germplasm revealed by simple sequence repeat markers. Am. J. Potato Res. 2009, 86, 38–48. [Google Scholar] [CrossRef]
- Yıldırım, A.; Kandemir, N.; Sönmezoğlu, Ö.A.; Güleç, T.E. Transferability of microsatellite markers among cool season cereals. Biotechnol. Biotechnol. Equip. 2009, 23, 1299–1302. [Google Scholar] [CrossRef] [Green Version]
- Favoretto, P.; Veasey, E.A.; Melo, P.C.T. Molecular characterization of potato cultivars using SSR markers. Hortic. Bras. 2011, 29, 542–547. [Google Scholar] [CrossRef] [Green Version]
- Provan, J.; Powell, W.; Hollingsworth, P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2001, 16, 142–147. [Google Scholar] [CrossRef]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Andersen, J.R.; Lübberstedt, T. Functional markers in plants. Trends Plant Sci. 2003, 8, 554–560. [Google Scholar] [CrossRef]
- Yadav, P.; Vaidya, E.; Rani, R.; Yadav, N.K.; Singh, B.K.; Rai, P.K.; Singh, D. Recent Perspective of Next Generation Sequencing: Applications in Molecular Plant Biology and Crop Improvement. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 435–449. [Google Scholar] [CrossRef]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001, 29, e25. [Google Scholar] [CrossRef] [Green Version]
- Wenzl, P.; Carling, J.; Kudrna, D.; Jaccoud, D.; Huttner, E.; Kleinhofs, A.; Kilian, A. Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. USA 2004, 101, 9915–9920. [Google Scholar] [CrossRef] [Green Version]
- Courtois, B.; Audebert, A.; Dardou, A.; Roques, S.; Herrera, T.G.; Droc, G.; Frouin, J.; Rouan, L.; Gozé, E.; Kilian, A.; et al. Genome-Wide Association Mapping of Root Traits in a Japonica Rice Panel. PLoS ONE 2013, 8, e78037. [Google Scholar] [CrossRef] [Green Version]
- Hahn, V.; Würschum, T. Molecular genetic characterization of Central European soybean breeding germplasm. Plant Breed. 2014, 133, 748–755. [Google Scholar] [CrossRef]
- Von Mark, V.C.; Kilian, A.; Dierig, D.A. Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS ONE 2013, 8, e64062. [Google Scholar]
- Raman, H.; Raman, R.; Coombes, N.; Song, J.; Prangnell, R.; Bandaranayake, C.; Tahira, R.; Sundaramoorthi, V.; Killian, A.; Meng, J.; et al. Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 2016, 39, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Dracatos, P.M.; Haghdoust, R.; Singh, R.P.; Huerta Espino, J.; Barnes, C.W.; Forrest, K.; Hayden, M.; Niks, R.E.; Park, R.F.; Singh, D. High-Density Mapping of Triple Rust Resistance in Barley Using DArT-Seq Markers. Front. Plant Sci. 2019, 10, 467. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ren, R.; Ray, R.; Xu, J.; Li, P.; Zhang, M.; Liu, G.; Yao, X.; Kilian, A. Genetic diversity and population structure of core watermelon (Citrullus lanatus) genotypes using DArTseq-based SNPs. Plant Genet. Resour. 2016, 14, 226–233. [Google Scholar] [CrossRef]
- Al-Beyroutiová, M.; Sabo, M.; Sleziak, P.; Dušinský, R.; Birčák, E.; Hauptvogel, P.; Kilian, A.; Švec, M. Evolutionary relationships in the genus Secale. Plant Syst. Evol. 2016, 302, 1083–1091. [Google Scholar] [CrossRef]
- Farahani, S.; Maleki, M.; Mehrabi, R.; Kanouni, H.; Scheben, A.; Batley, J.; Talebi, R. Whole Genome Diversity, Population Structure, and Linkage Disequilibrium Analysis of Chickpea (Cicer arietinum L.) Genotypes Using Genome-Wide DArTseq-Based SNP Markers. Genes 2019, 10, 676. [Google Scholar] [CrossRef] [Green Version]
- Hassani, S.M.R.; Talebi, R.; Pourdad, S.S.; Naji, A.M.; Fayaz, F. In-depth genome diversity, population structure and linkage disequilibrium analysis of worldwide diverse safflower (Carthamus tinctorius L.) accessions using NGS data generated by DArTseq technology. Mol. Biol. Rep. 2020, 47, 2123–2135. [Google Scholar] [CrossRef]
- Śliwka, J.; Jakuczun, H.; Chmielarz, M.; Hara-Skrzypiec, A.; Tomczyńska, I.; Kilian, A.; Zimnoch-Guzowska, E. A resistance gene against potato late blight originating from Solanum × michoacanum maps to potato chromosome VII. TAG. Theor. Appl. Genet. 2011, 124, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorizzo, M.; Gao, L.; Mann, H.; Traini, A.; Chiusano, M.L.; Kilian, A.; Aversano, R.; Carputo, D.; Bradeen, J.M. A DArT marker-based linkage map for wild potato Solanum bulbocastanum facilitates structural comparisons between Solanum A and B genomes. BMC Genet. 2014, 15, 123. [Google Scholar] [CrossRef] [Green Version]
- Nachtigall, M.; Konig, J.; Thieme, R. Mapping of a novel, major late blight resistance locus in the diploid (1EBN) Mexican Solanum pinnatisectum Dunal on chromosome VII. Plant Breed. 2018, 137, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Dracatos, P.M.; Zhang, P.; Park, R.F.; McIntosh, R.A.; Wellings, C.R. Complementary resistance genes in wheat selection ‘Avocet R’confer resistance to stripe rust. Theor. Appl. Genet. 2016, 129, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Mogga, M.; Sibiya, J.; Shimelis, H.; Lamo, J.; Yao, N. Diversity analysis and genome-wide association studies of grain shape and eating quality traits in rice (Oryza sativa L.) using DArT markers. PLoS ONE 2018, 13, e0198012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atencio, M. Diversidad en Variedades Andinas de Papa Solanum Tuberosum ssp. Andigena Evaluada con Microsatélites [Diversity on Andean Potato Varieties Solanum tuberosum ssp. Andigena Evaluated with Microsatellites]. Master’s Thesis, Facultad de Ciencias Agrarias, Universidad Nacional de Mar del Plata, Mar del Plata, Argentina, 2011. [Google Scholar]
- GCDT. Background on the development of the ‘Global Strategy for the Ex situ Conservation of Potato’. 2006. Available online: https://www.croptrust.org/wp-content/uploads/2014/12/Potato-Strategy-FINAL-30Jan07.pdf (accessed on 5 April 2019).
- Okada, K.A. Exploration, conservation and evaluation of potato germplasm in Argentina. Potato Res. 1976, 19, 263–269. [Google Scholar] [CrossRef]
- Okada, K.A. Collection and taxonomy of Argentine wild species (tuber-bearing Solanum). In Report of the Planning Conference on the Exploration, Taxonomy and Maintenance of Potato Germplasm, 15–19 October 1979; CIP: Lima, Peru, 1979; pp. 98–113. [Google Scholar]
- Okada, K.A.; Clausen, A.M. Collecting potatoes in northwestern Argentina. Am. Potato J. 1983, 64, 301–305. [Google Scholar]
- Clausen, A.M. Collecting indigenous potato varieties in Northwest Argentina. Plant Genet. Resour. Newsl. 1989, 80, 38–39. [Google Scholar]
- Clausen, A.M.; Colavita, M.; Butzonitch, I.; Carranza, A.V. A potato collecting expedition in the province of Jujuy, Argentina and disease indexing of virus and fungus pathogens in Andean cultivars. Genet. Resour. Crop. Evol. 2005, 52, 1099–1109. [Google Scholar] [CrossRef]
- Haymes, H. Mini-prep method suitable for plant breeding program. Plant Mol. Biol. Rep. 1996, 14, 280–284. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.H.; Guo, H.; Wang, X.; Kim, C.H.; Paterson, A.H. SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genom. 2014, 15, 162. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. PHYLIP-phylogeny inference package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3, an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Batley, J.; Barker, G.; O’Sullivan, H.; Edwards, K.J.; Edwards, D. Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol. 2003, 132, 84–91. [Google Scholar] [CrossRef] [Green Version]
- Pootakham, W.; Jomchai, N.; Ruang-Areerate, P.; Shearman, J.R.; Sonthirod, C.; Sangsrakru, D.; Tragoonrung, S.; Tangphatsornruang, S. Genome-wide SNP discovery and identification of QTL associated with agronomic traits in oil palm using genotyping-by-sequencing (GBS). Genomics 2015, 105, 288–295. [Google Scholar] [CrossRef] [Green Version]
- Shearman, J.R.; Sangsrakru, D.; Jomchai, N.; Ruang-Areerate, P.; Sonthirod, C.; Naktang, C.; Theerawattanasuk, K.; Tragoonrung, S.; Tangphatsornruang, S. SNP identification from RNA sequencing and linkage map construction of rubber tree for anchoring the draft genome. PLoS ONE 2015, 10, e0121961. [Google Scholar] [CrossRef]
- Soorni, A.; Fatahi, R.; Haak, D.C.; Salami, S.A.; Bombarely, A. Assessment of Genetic Diversity and Population Structure in Iranian Cannabis Germplasm. Nat. Sci. Rep. 2017, 7, 15668. [Google Scholar] [CrossRef]
- Hayashi, K.; Hashimoto, N.; Daigen, M.; Ashikawa, I. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor. Appl. Genet. 2004, 108, 1212–1220. [Google Scholar] [CrossRef]
- Picoult-Newberg, L.; Ideker, T.E.; Pohl, M.G.; Taylor, S.L.; Donaldson, M.A.; Nickerson, D.A.; Boyce-Jacino, M. Mining SNPs from EST databases. Genome Res. 1999, 9, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, S.; Iseli, I.; Bucher, P.; Antonarakis, S.E.; Scott, H.S. A cSNP map and database for human chromosome 21. Genome Res. 2011, 11, 300. [Google Scholar] [CrossRef]
- Ochoa, C.M. The Potatoes of South America: Bolivia; Cambridge University Press: Cambridge, UK, 1990. [Google Scholar]
- Hirsch, C.N.; Hirsch, C.; Felcher, K.; Coombs, J.; Zarka, D.; Van Deynze, A.; De Jong, W.; Veilleux, R.E.; Jansky, S.; Bethke, P.; et al. Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 Genes Genomes Genet. 2013, 3, 1003–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bali, S.; Robinson, B.R.; Sathuvalli, V.; Bamberg, J.; Goyer, A. Single Nucleotide Polymorphism (SNP) markers associated with high folate content in wild potato species. PLoS ONE 2018, 13, e0193415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Colman, S. Contribución Alélica de Genes Candidatos al Endulzamiento Inducido por Frío en Papa Solanum tuberosum L. [Candidate Genes Allelic Contribution to Cold Sweetening Induce on Potato Solanum tuberosum L.]. Ph.D. Thesis, Facultad de Ciencias Exactas y Naturales, Universidad Nacional de Mar del Plata, Mar del Plata, Argentina, 2014. [Google Scholar]
- Wang, Y.; Rashid, M.A.R.; Li, X.; Yao, C.; Lu, L.; Bai, J.; Li, Y.; Xu, N.; Yang, Q.; Zhang, L.; et al. Collection and Evaluation of Genetic Diversity and Population Structure of Potato Landraces and Varieties in China. Front. Plant Sci. 2019, 10, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Group (Atencio, 2011) | Accession Code | Study Code | Landrace | Location (Province, Department, Locality) |
|---|---|---|---|---|
| 1 | CCS 1327 | 28 | Bayista | Jujuy, Cochinoca, Rachaite |
| 1 | CL 621 | 23 | Chorcoyeña | Salta, Santa Victoria, Nazareno |
| 1 | CL 516 | 44 | Chorcoyeña | Salta, Santa Victoria, Chorro |
| 1 | CCS 1205 | 21 | Churqueña | Jujuy, Humahuaca, Varas |
| 1 | CCS 1378 | 90 | Churqueña negra | Jujuy, Tumbaya, Patacal |
| 1 | CCS 1224 | 60 | Collareja | Jujuy, Humahuaca, Coctaca |
| 1 | CL 634 | 67 | Collareja | Salta, Santa Victoria, Arpero |
| 1 | CL 636 | 68 | Collareja | Salta, Santa Victoria, Abra Colorada |
| 1 | CS 1432 | 3 | Collareja or cuarentona overa | Jujuy, General Belgrano, Cuevas |
| 1 | CCS1381 | 29 | Runa | Jujuy, Tumbaya, Patacal |
| 1 | CL 650 | 45 | Runa | Salta, Santa Victoria, Poscaya |
| 1 | CL 576 | 46 | Runa | Salta, Santa Victoria, Lizoite |
| 1 | CL 641 | 49 | Runa | Salta, Santa Victoria, Poscaya |
| 1 | CL 708 | 56 | Runa | Salta, Iruya, Colanzulí |
| 1 | CL 489 | 63 | Runa | Salta, Santa Victoria, Rodeopampa |
| 1 | CL 739 | 70 | Runa | Jujuy, Humahuaca, Chaupi Rodero |
| 1 | CL 750 | 71 | Runa | Jujuy, Humahuaca, Chaupi Rodero |
| 1 | CCS 1218 | 84 | Runa | Jujuy, Humahuaca, Ocumazo |
| 1 | CCS 1340 | 116 | Yaguana | Jujuy, Susques, Sala |
| 1 | CL 641 | 144 | Runa | Salta, Santa Victoria, Poscaya |
| 2 | LC 348 | 62 | Imilla Negra | Jujuy, Humahuaca, Huachichocana |
| 2 | CL 631 | 40 | Allo | Salta, Iruya, Campo Carreras |
| 2 | CCS 1227 | 109 | Azul or Sallama | Jujuy, Humahuaca, Coctaca |
| 2 | CCS 1196 | 80 | Azul overa | Jujuy, Humahuaca, Palca de Aparzo |
| 2 | CL 815 | 77 | Boliviana | Jujuy, Valle Grande, Santa Ana |
| 2 | CCS 1201 | 14 | Condorilla | Jujuy, Humahuaca, Varas |
| 2 | CCS 1384 | 41 | Corbatilla | Jujuy, Tumbaya, Patacal |
| 2 | CCS 1330 | 6 | Moradita | Jujuy, Cochinoca, Rachaite |
| 2 | CCS 1307 | 8 | Moradita | Jujuy, Santa Catalina, Cabreria |
| 2 | CCS 1374 | 39 | Moradita | Jujuy, Cochinoca, Agua Castilla |
| 2 | CCS 1172 | 4 | Moradita redonda | Jujuy, Tilcara, Casa Colorada |
| 2 | CL 783 | 98 | Navecilla | Jujuy, Valle Grande, Santa Ana |
| 2 | CL 820 | 57 | Negra Redonda | Jujuy, Valle Grande, Santa Ana |
| 2 | CCS 1305 | 47 | Ojosa | Jujuy, Santa Catalina, Casir |
| 2 | CCS 1257 | 86 | Ojosa | Jujuy, Rinconada, Rinconada |
| 2 | CCS 1366 | 27 | Overa | Jujuy, Tumbaya, El Moreno |
| 2 | CL 748 | 35 | Overa | Jujuy, Humahuaca, Chaupi Rodero |
| 2 | CL 790 | 52 | Overa | Jujuy, Valle Grande, Santa Ana |
| 2 | CL 832 | 79 | Abajeña overa | Jujuy, Valle Grande, Santa Ana |
| 2 | CL 793 | 74 | Sallama | Jujuy, Valle Grande, Santa Ana |
| 2 | CL 804 | 139 | Sallama | Jujuy, Valle Grande, Santa Ana |
| 2 | CL 769 | 32 | Sallama Grande | Jujuy, Valle Grande, Santa Ana |
| 2 | CCS 1284 | 26 | Sani | Jujuy, Yavi |
| 2 | CCS 1303 | 37 | Yuruma | Jujuy, Santa Catalina, Casira |
| 2 | CCS 1385 | 42 | Moradita | Jujuy, Tumbaya, Patacal |
| 3 | CL 835 | 51 | Airampía | Jujuy, Valle Grande, Santa Ana |
| 3 | CL 836 | 54 | Airampía | Jujuy, Valle Grande, Santa Ana |
| 3 | CL 528 | 64 | Colorada | Salta, Santa Victoria, Chorro |
| 3 | CCS1221 | 102 | Colorada | Jujuy, Humahuaca, Coctaca |
| 3 | CL 508 | 123 | Colorada | Salta, Santa Victoria, Chorro |
| 3 | CCS 1349 | 12 | Coloradita | Jujuy, Tumbaya, El Angosto |
| 3 | CCS 1184 | 108 | Colorana or Señorita | Jujuy, Humahuaca, Aparzo |
| 3 | CL 821 | 78 | Cuarentilla Toscra | Jujuy, Valle Grande, Santa Ana |
| 3 | CS 1430 | 16 | Cuarentona | Jujuy, General Belgrano, Cuevas |
| 3 | CL 728 | 31 | Cuarentona | Salta, Iruya, Colanzulí |
| 3 | CCS 1166 | 9 | Cuarentona colorada | Jujuy, Tilcara, Casa Colorada |
| 3 | CS 1414 | 94 | Cuarentona morada | Jujuy, General Belgrano, Papachacra |
| 3 | CS 1425 | 122 | Cuarentona oquecha | Jujuy, General Belgrano, Papachacra |
| 3 | CCS 1353 | 117 | Cuarentona Redonda | Jujuy, Tumbaya, El Angosto |
| 3 | CS 1416 | 120 | Cuella | Jujuy, General Belgrano, Papachacra |
| 3 | CCS 1288 | 15 | Desiree | Jujuy, Santa Catalina, Cieneguillas |
| 3 | CL 712 | 69 | Huareña | Salta, Iruya, Colanzulí |
| 3 | CCS 1170 | 53 | Ojos colorados | Jujuy, Tilcara, Casa Colorada |
| 3 | CS 1402 | 91 | Ojos colorados | Jujuy, Tumbaya, Carcel |
| 3 | CCS 1383 | 34 | Pera or señorita | Jujuy, Tumbaya, Patacal |
| 3 | CCS 1321 | 89 | Rosada | Jujuy, Cochinoca, Agua Caliente |
| 3 | CL 658 | 17 | Santa María | Jujuy, Yavi, Yavi |
| 3 | LC 335 | 97 | Tonca | Jujuy, Humahuaca, Patacal |
| 3 | CL 482 | 33 | Rosada | Salta, Santa Victoria, Rodeopampa |
| 3 | CCS 1255 | 7 | Desiree | Jujuy, Rinconada, Rinconada |
| 3 | CCS 1323 | 36 | Colorada | Jujuy, Cochinoca, Agua Caliente |
| 4 | CL 849 | 99 | Balcacha | Salta, Rosario de Lerma, El Gólgota |
| 4 | CCS 1350 | 1 | Blanca | Jujuy, Tumbaya, El Angosto |
| 4 | CS 1419 | 10 | Blanca | Jujuy, General Belgrano, Papachacra |
| 4 | CCS 1310 | 115 | Blanca alargada | Jujuy, Santa Catalina, Cabreria |
| 4 | CCS 1309 | 58 | Blanca redonda | Jujuy, Santa Catalina, Cabreria |
| 4 | CCS 1251 | 5 | Chacarera | Jujuy, Cochinoca, Cochinoca |
| 4 | CCS 1371 | 20 | Chacarera | Jujuy, Cochinoca, Quebraleña |
| 4 | CS 1418 | 2 | Chaqueña | Jujuy, General Belgrano, Papachacra |
| 4 | CS 1408 | 92 | Chaqueña overa | Jujuy, General Belgrano, Papachacra |
| 4 | CCS 1300 | 88 | Holandesa | Jujuy, Santa Catalina, Casira |
| 4 | CCS 1209 | 83 | Luqui | Jujuy, Humahuaca, Chorcan |
| 4 | CCS 1299 | 100 | Malgacha | Jujuy, Santa Catalina, Casira |
| 4 | CCS 1200 | 81 | Papa oca | Jujuy, Humahuaca, Varas |
| 4 | CCS 1206 | 82 | Papa oca | Jujuy, Humahuaca, Varas |
| 4 | CL 548 | 48 | Papa palta | Salta, Santa Victoria, Trigohuaico |
| 4 | CS 1413 | 93 | Papa vallista | Jujuy, General Belgrano, Papachacra |
| 4 | CCS 1185 | 13 | Tuni | Jujuy, Humahuaca, Aparzo |
| 4 | CCS 1199 | 11 | Tuni blanca | Jujuy, Humahuaca, Palca de Aparzo |
| 4 | CCS 1247 | 22 | Tuni blanca | Jujuy, Cochinoca, Ojo de Agua |
| 4 | CCS 1375 | 101 | Tuni Blanca | Jujuy, Tumbaya, Tumbaya |
| 4 | CCS 1393 | 119 | Tuni morada | Jujuy, Tumbaya, Cieneguillas |
| 4 | CL 782 | 73 | Tuni rosilla | Jujuy, Valle Grande, Santa Ana |
| 4 | CCS 1271 | 25 | Blanca | Jujuy, Santa Catalina, Morco Esquina |
| 4 | Cl 752B | 72 | Blanca | Jujuy, Valle Grande, Santa Ana |
| 4 | CL 814A | 75 | Holandesa colorada | Jujuy, Valle Grande, Santa Ana |
| Berta | 104 | Azul | Provided by Jujuy National University | |
| Berta | 105 | Santa María | Provided by Jujuy National University | |
| Berta | 106 | Santa María pulpa blanca | Provided by Jujuy National University | |
| 70B | Guacha potato | |||
| 89A | Guacha potato | |||
| 89B | Guacha potato | |||
| DM | Potato reference genome | |||
| IN1 | Imilla Negra | Provided by Cauqueva | ||
| IN2 | Imilla Negra | Provided by Cauqueva | ||
| IN3 | Imilla Negra | Provided by Cauqueva | ||
| IN4 | Imilla Negra | Provided by Cauqueva | ||
| IN5 | Imilla Negra | Provided by Cauqueva | ||
| IN6 | Imilla Negra | Provided by Cauqueva | ||
| IN7 | Imilla Negra | Provided by Cauqueva | ||
| BINTJE | Bintje | Commercial variety | ||
| PAMPEANA-INTA | Pampeana-Inta | Commercial variety | ||
| SPUNTA | Spunta | Commercial variety | ||
| SPUNTA2 | Spunta | Commercial variety |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sucar, S.; Carboni, M.F.; Rey Burusco, M.F.; Castellote, M.A.; Massa, G.A.; Monte, M.N.; Feingold, S.E. Assessment of Genetic Diversity and Relatedness in an Andean Potato Collection from Argentina by High-Density Genotyping. Horticulturae 2022, 8, 54. https://doi.org/10.3390/horticulturae8010054
Sucar S, Carboni MF, Rey Burusco MF, Castellote MA, Massa GA, Monte MN, Feingold SE. Assessment of Genetic Diversity and Relatedness in an Andean Potato Collection from Argentina by High-Density Genotyping. Horticulturae. 2022; 8(1):54. https://doi.org/10.3390/horticulturae8010054
Chicago/Turabian StyleSucar, Sofía, Martín Federico Carboni, María Florencia Rey Burusco, Martín Alfredo Castellote, Gabriela Alejandra Massa, Marcelo Nicolás Monte, and Sergio Enrique Feingold. 2022. "Assessment of Genetic Diversity and Relatedness in an Andean Potato Collection from Argentina by High-Density Genotyping" Horticulturae 8, no. 1: 54. https://doi.org/10.3390/horticulturae8010054
APA StyleSucar, S., Carboni, M. F., Rey Burusco, M. F., Castellote, M. A., Massa, G. A., Monte, M. N., & Feingold, S. E. (2022). Assessment of Genetic Diversity and Relatedness in an Andean Potato Collection from Argentina by High-Density Genotyping. Horticulturae, 8(1), 54. https://doi.org/10.3390/horticulturae8010054