1. Introduction
Coatings that are cured using ultraviolet light (“UV-curables”) find use in a wide range of applications as they can be rapidly cured without the evolution of unpleasant or noxious gases. UV-curables contain a certain amount of photoinitiator that strongly absorbs in the UV range. When this photoinitiator absorbs UV light, radicals are formed that drive the formation of crosslinks between monomers to form a polymeric coating [
1]. Greater UV absorbance, either from more efficient photoinitiators or by more intense UV light, results in more radicals that, in turn, promote curing of the coating [
2,
3]. One aspect of these reactions must frequently be addressed: atmospheric oxygen can interfere with radical-driven crosslinking reactions at the coating surface. In our experiments, oxygen reacts rapidly with radicals produced during photoinitiation to produce peroxyl radicals that are slow to react with double bonds in monomers, thus retarding the rate of polymerization [
4]. Sometimes curing can be performed in an oxygen-free atmosphere, minimizing this interference. Alternatively, when atmospheric regulation is impractical, antioxidant scavengers can be added to the coating mixture to mitigate the effects of (atmospheric) oxygen radicals, allowing the main crosslinking reaction to proceed [
5]. One efficient scavenger is triphenylphosphine (PPh
3) which, in our experiments, reacts with a peroxyl radical to form triphenylphosphine oxide and a reactive radical monomer than can participate in chain propagation [
4]. Details of these curing processes are described elsewhere [
6].
The extent of crosslinking in UV-curable coatings can be quantified in several ways. Fourier-transform infrared spectroscopy (FTIR) can show the conversion of chemical bonds from monomer to polymer, indicating the extent of crosslinking [
1]. Although widely used, surface FTIR (attenuated total reflection, or ATR-FTIR) is typically limited to a very shallow depth of the coating—the exact depth is quantitatively dependent on indices of refraction and on the incident angle of the IR light, but typical experimental values range from 0.6 to 2 μm [
7]. (Certain research applications may use infrared spectroscopy in a transmission mode, but that approach is inappropriate for coatings applied to IR-opaque substrates.) Depending on the sample, the penetration depth of ATR-FTIR may be appropriate to measure curing at the surface of the coating, but may not represent the curing throughout the entire thickness of the coating. Other spectroscopic techniques, including Raman spectroscopy, ultimately face the same question of penetration depth. Penetration depth is, of course, also affected by the opacity of the sample at the particular wavelength of light used in the experiment.
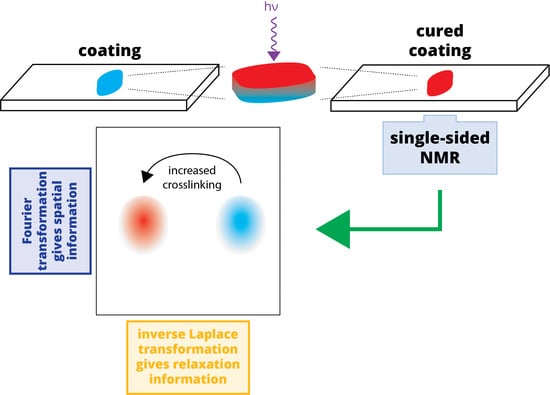
For many applications, nuclear magnetic resonance (NMR) offers an attractive alternative to traditional optical spectroscopy for quantifying crosslinking in polymeric films, including UV-curable coatings. While traditional, high-field NMR typically requires samples to be in a 5-mm diameter NMR tube that is inserted into the bore of the superconducting magnet, single-sided NMR devices can directly probe materials at surfaces without sampling or sample preparation [
8,
9]. These devices, while unable to record chemical-shift–resolved spectra, can measure relaxation of nuclear spin states, which is directly affected by the degree of crosslinking [
10]. In this regard, single-sided magnets perform similarly to optical spectroscopy, albeit with much lower sensitivity. However, the design of these magnets imparts a fixed and strong magnetic field gradient that, in analogy to clinical MRI, can be used in an imaging modality via Fourier transformation. In this way, the extent of curing can be probed in a spatially selective manner throughout the entire thickness of the sample, even for samples over 100 μm thick, with a spatial resolution on the order of a few microns [
11].
In this report, we describe the use of single-sided magnets to investigate the extent of crosslinking in a UV-curable polymer system comprising poly(ethylene glycol) diacrylate (PEGDA), in a spatially sensitive manner. The effects of the scavenger molecule PPh
3, the UV intensity, and the thickness of the coating on the extent of curing are reported. In addition, we report the use of sequential Fourier and inverse Laplace transformations to visualize the relaxation cross-section of a sample that describes the extent of crosslinking throughout the sample thickness. Spatially selective NMR measurements of coatings have only been lightly reported in the literature [
12], and those reports have not leveraged Fourier transformations of the NMR data to optimize spatial resolution. These steps improve the utility of single-sided NMR in investigating (polymeric) films via their spatially variant relaxation times.
2. Results
NMR data collected using single-sided magnets cannot capture chemical shift information. When using these devices, typical experiments are made with a single excitation pulse followed by a train of refocusing pulses; interleaved between refocusing pulses are acquisition windows that each capture the refocused magnetization “echo”: a record of the signal intensity that no longer contains chemical shift information. This Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence is crucial to the success of single-sided NMR, as its refocusing sustains measurable signals long enough to be successfully recorded in a highly inhomogeneous magnetic field [
13,
14]. Consequently, the CPMG pulse sequence can be used to evaluate time-dependent signal attenuation that is ultimately related to physical properties of materials. In a CPMG measurement, each echo, like traditional free-induction decay (FID) signals, is made of some number of complex data points each measured over some dwell time. In our data processing, each echo is individually subjected to Fourier transformation, converting the time domain echo signal to a frequency domain signal that, because of the permanent magnetic field gradient, is proportional to a spatial domain signal. Once every echo has been transformed into the spatial domain, the signal decay (over the many successive echoes) at each position is subjected to an inverse Laplace transformation (ILT), the result of which is a relaxation “spectrum” at each position. An ILT produces a spectrum showing the intensity of various relaxation rates that, when added, best represent the overall signal attenuation in the signal: smaller
T2 relaxation times (to the left of the ILT spectrum) indicate greater relaxation which, in turn, suggests more crosslinking [
15]. The final product of this data processing approach is a relaxation cross-section (position,
z, vs.
T2) that shows differences in local crosslinking at different depths in the sample. The details of ILTs are described in detail elsewhere [
16], and the entire data processing procedure is outlined in
Figure 1.
The relaxation cross-section (or
z-
T2 map) highlights the
T2 relaxation values at different positions within the sample. As an example,
Figure 2 shows a
z-
T2 map of a PEGDA sample that was not spun, and remains relatively thick (~80 μm). The UV light penetrates only the top ~20 μm of this thick sample (the top of the sample is in the negative
z direction), which becomes crosslinked (smaller
T2, around 10
−3 s), while the lower ~60 μm of the coating (away from the UV light, towards the positive
z direction) remain less crosslinked (larger
T2, around 10
−2 s). This method of data presentation highlights spatial variations in relaxation times and is used throughout this report.
Figure 3 highlights the correspondence between ATR-FTIR and
T2 measurements of crosslinking in the PEGDA network. The IR spectra are normalized to the peak at 1726 cm
−1, corresponding to the acrylic carbonyl group; these carbonyl groups are largely unaffected during curing and their IR intensities should remain fixed. In contrast, the terminal CH=CH
2 bonds on the PEGDA monomers produce a peak at 1410 cm
−1 [
17,
18,
19,
20]. A decrease in the intensity of the 1410 cm
−1 peak when the sample is exposed to UV light indicates curing. Simultaneously, the measured
T2 value decreases after the sample has been exposed to UV irradiation. Crosslinking is associated with reduced
T2 values, as reported elsewhere [
15].
When radical-driven curing of PEGDA polymers is performed in the presence of atmospheric oxygen, the oxygen can form radicals that interfere with crosslinking in the polymer network [
4]. Several methods can be employed to circumvent this, including curing in an anaerobic environment (requiring engineering controls) or adding a scavenger molecule (PPh
3 in our samples) that mitigates the deleterious effects of oxygen [
4,
5].
Figure 4 shows
z-
T2 maps of several samples that are cured in (an oxygen-containing) lab atmosphere. The leftmost column of
Figure 4 (
Figure 4a–f) lacks PPh
3. Their
z-
T2 maps highlight that there is no change in
T2 (i.e., no crosslinking) in the samples regardless of the UV power deposited into the sample. Furthermore, there is little evidence of a curing gradient throughout the thickness of the sample (a spatially dependent
T2), suggesting that the oxygen-driven inhibition effect takes place, as expected, at the sample/air interface. Possible exceptions to this are in
Figure 4e,f, in which “tails” with shorter
T2s appear near the bottom (positive
z direction, near the glass interface) of the sample. In these samples, exposed to the highest UV power, it is possible that some curing occurs ~15 μm below the sample surface. This suggests that, although atmospheric oxygen inhibits curing at the coating surface, some crosslinking may still occur in regions of the sample inaccessible to oxygen provided there is sufficient UV power.
The second column of
Figure 4 (
Figure 4g–l) shows the effect of increasing amounts of the scavenger PPh
3, from 0–5%
w/
w, on crosslinking in the PEGDA sample. At low concentrations of PPh
3, the sample retains a large
T2 value (less crosslinking), and as PPh
3 increases,
T2 decreases, indicating more crosslinking. Using these data, we elected to use 1% PPh
3 (
Figure 4j) as a standard sample to evaluate the extent of crosslinking as a function of UV power. This power-dependent series (0–100% UV power, 1% PPh
3) are shown in the rightmost column of
Figure 4 (
Figure 4m–r). As expected, measured
T2 values decrease with increasing UV power; again, this is consistent with increased crosslinking in the sample.
3. Discussion
Single-sided NMR is highly conducive to in situ measurements of coatings on surfaces. Along with optical methods (e.g., ATR-FTIR and Raman), NMR can probe the extent of crosslinking within a sample; in magnetic resonance contexts, this is most easily measured as a change in the spin-spin relaxation time
T2. However, NMR offers a much larger spatial range (in the depth dimension) for measurements than do non-confocal optical techniques. In this regard, we observe a crosslinking “gradient” throughout the thickness of a particularly thick UV-cured coating sample (see
Figure 2). NMR is successful in evaluating crosslinking differences that are due to both different UV intensities and to different chemical compositions (different amounts of scavenger PPh
3 in our case). In other words, single-sided NMR is insensitive to the cause of differences in crosslinking, which may prove useful where in situ analyses are intended only to measure the extent of crosslinking.
Despite its ability to probe thick samples, single-sided NMR measurements show poor spatial resolution relative to many optical techniques, both laterally and in the depth dimension. Single-sided NMR devices use surface coils that are typically several square centimeters in area, meaning that relaxation measurements reflect a lateral spatial average over that area. This is less problematic for homogeneous materials (such as coatings), but needs to be considered during experimental design. The spatial resolution in the depth dimension is fundamentally related to the magnetic field gradient and the acquisition parameters: strong field gradients and large acquisition times (both possible with single-sided devices) result in high spatial resolution. Practically, the coplanarity of the sample with the magnet’s “sweet spot” restricts the resolution in the depth dimension: less-than-perfect coplanarity between the sample and the sweet spot will blur the resolution of the measurement. Generally, the depth resolution of single-sided NMR measurements is on the order of 10 μm, though careful calibration and experimental setup can improve this several-fold [
11].
Simple future experiments with single-sided NMR extends this work to coatings of other compositions, including different effective molecular weights of PEGDA. A more sophisticated approach may be to exploit dipolar coupling in crosslinked materials to more quantitatively probe crosslinking in a material. This method has been demonstrated elsewhere on elastomers [
22,
23] and may circumvent some of the limitations of
T2 determinations from CPMG data, in particular a strong dependence on both equipment-specific and user-selected experimental parameters. Furthermore, single-sided NMR may be particularly well suited to evaluating the extent of crosslinking within interpenetrating polymer networks (IPNs) [
24]; these coatings consist of two or more phases that in tandem produce specific material properties.
4. Materials and Methods
4.1. Samples
Samples comprised different ratios of poly(ethylene glycol) diacrylate (Mn = 700 g mol−1), photoinitiator 1-hydroxycyclohexyl phenyl ketone (HPCK, 2% w/w), and triphenylphosphine (PPh3). All chemicals were used as received without further purification (Sigma-Aldrich, St. Louis, MO, USA). HCPK and PPh3 were dissolved into acetone prior to being added to the sample. The amount of PPh3 varied per experiment from 0% to 5% by weight with the total amount of acetone fixed at 7% w/v. Slides were prepared with a piece of labeling tape in the middle with a ~1 cm diameter circle cut out in the tape; this tape was designed to contain the sample in a region that could be uniformly cured by the UV gun and that would fit in the sensitive region of the NMR magnet. Ten microliters of the sample were placed on a glass slide in the center of the circle. Most slides were spun at a rate of ~3000 rpm for 30 s using a 3D-printed spin coater driven by a computer hard drive motor. (One sample slide was intentionally left thick and was not spun.) While spinning, the sample, at a distance of ~1.4 cm from the UV source, was cured using a ThorLabs CS2010 LED UV curing system (ThorLabs, Inc., Ann Arbor, MI, USA). At 100%, the UV system outputs 185 mW cm−2 at 365 nm (instrument-internal calibration). The power of the UV system was adjusted to 20%, 40%, 60%, 80%, and 100% of the maximum power—the output varied linearly with nominal power. After spinning and curing, samples had a thickness of ~20 μm, determined using a Profilm3D surface profiler (Filmetrics, Inc.; San Diego, CA, USA).
4.2. NMR Measurements
All NMR measurements were made with a PM5 NMR-MOUSE (
B0 = 0.4 T) operated by a Kea2 spectrometer (Magritek, Ltd., Wellington, New Zealand). An initial CPMG [
13,
14] sequence was run on each slide to center the magnet to find the most signal. The
B1 frequency was adjusted by a maximum of 50 kHz (~50 μm) to center the maximum signal in the acquisition window. (The rf coil has a bandwidth of ~350 kHz, so these adjustments do not substantially affect the coil sensitivity). This short CPMG had 512 scans, a 1 μs dwell time and 128 complex points. Subsequently, a final CPMG was run for data collection with 4096 scans, 10 μs dwell time, and 32 complex points with all other parameters staying constant between slides. NMR measurements were taken directly after curing to minimize outside factors, such as extra exposure to UV light.
4.3. FTIR Measurements
FTIR data were collected using a Digital Labs FTS 7000 ATR-FTIR (Varian, Inc., Palo Alto, CA, USA). The samples were made of PEGDA with 2% HCPK and 2% PPh3 (w/w) and were prepared in the same manner as those for NMR data collection. A blank microscope slide was used for the background scan; all measurements were made from 600 cm−1 to 2000 cm−1 with 1 cm−1 resolution for 64 scans.
4.4. Data Processing
All data processing was performed in Matlab (Mathworks, Inc., Natick, MA, USA) using custom processing scripts. These scripts for importing and Fourier transforming the NMR data were made in house and are available at the institutional repository of the corresponding author. Inverse Laplace transformation was also performed in Matlab using scripts provided by Petrik Galvosas (University of Wellington, New Zealand) [
25].