Enhancement and Segmentation Workflow for the Developing Zebrafish Vasculature †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry
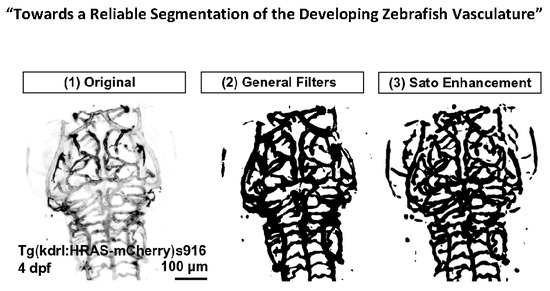
- Tg(kdrl:HRAS-mCherry)s916 [14]: membrane tagged mCherry under the endothelial-specific kdrl promotor;
- Tg(fli1a:eGFP)y1 [15]: cytosolic eGFP under the pan-endothelial fli1a promotor;
- Tg(fli1a:LifeAct-mClover)sh467 (will be described elsewhere): endothelial filamentous actin tagged with mClover fluorophore under the pan-endothelial fli1a promotor.
2.2. Image Acquisition
- Images of 3 days post fertilization (dpf) Tg(fli1a:eGFP)y1, Tg(kdrl:HRAS-mCherry)s916, and Tg(fli1a:Lifeact-mClover)sh467 for CNR quantification;
- Images of 2-to-5 dpf Tg(kdrl:HRAS-mCherry)s916 for assessment of CNR, vascular enhancement, segmentation, and segmentation robustness;
- Time-lapse acquisitions with 200 cycles over 10 minutes (min) were performed with 3 s time intervals in 3 dpf Tg(kdrl:HRAS-mCherry)s916 and Tg(fli1a:eGFP)y1 to assess extent of motion and test the motion correction approach.
2.3. Data Analysis
2.3.1. Contrast-to-Noise Ratio (CNR)
2.3.2. Image Pre-Processing
- Tubular Filtering (TF): Fiji Tubeness Plugin, based on Sato [16], and implemented by Mark Longair, Stephan Preibisch and Johannes Schindelin [17]. The effect of varying the TF scale parameter (sigma) on the vascular double-peak intensity distribution was evaluated for the following values: 5.3424 (16 px), 8.0232 (24 px), 9.3604 (28 px), 10.6848 (32 px), 15.359 (46 px), and 23.718 (69 px).
2.3.3. Image Segmentation and Total Volume Measurement
- (i)
- Global Otsu thresholding using 16-bit images [22].
- (ii)
- k-means clustering using 16-bit images [23], initialized using the default 48 randomized seeds automatically placed by the k-means++ algorithm [24]. Variation of seed number was not found to improve segmentation results in the tested range of 20–70 seeds. Additional parameters were set as follows: 0.0001 cluster centre tolerance and interpretation as 3D stack. Detection of four clusters was chosen as this was found to deliver reliable results, especially after TF (one background cluster and three vessel clusters with varying brightness).
- (iii)
- (iv)
- Level set [27] using 8-bit images (to achieve acceptable processing times) with 50 user-specified vascular seeds, using the “Fast Marching” option with a distance threshold of zero and user-selected image-specific grey value thresholds.
2.4. Statistics and Data Representation
3. Results and Discussion
3.1. Image Pre-Processing
3.1.1. Assessment of Image Quality by CNR Quantification
3.1.2. Correction of Motion Artefacts
3.2. Vascular Enhancement and Segmentation
3.2.1. Vascular Enhancement
3.2.2. Segmentation Approaches
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACeV | anterior cerebral vein |
| BA | basal artery |
| CNR | contrast-to-noise ratio |
| CoV | coefficient of variance |
| DA | dorsal aorta |
| DLAV | dorsal longitudinal vein |
| dpf | days post fertilization |
| GF | general filtering |
| hpf | hours post fertilization |
| H | Hours |
| ISV | intersomitic vessel |
| LP | laser power |
| LSFM | light sheet fluorescence microscopy |
| min | Minutes |
| MIP | Maximum intensity projection |
| MMCtA | middle mesencephalic central artery |
| PCeV | posterior cerebral vein |
| PCS | posterior communicating segment |
| PMBC | primordial midbrain channel |
| PrA | prosencephalic artery |
| ROI | region of interest |
| s | Seconds |
| SRM | statistical region merging |
| TF | tubular filtering |
References
- Gut, P.; Reischauer, S.; Stainier, D.Y.R.; Arnaout, R. Little Fish, Big Data: Zebrafish as a Model for Cardiovascular and Metabolic Disease. Physiol. Rev. 2017, 97, 889–938. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.R.; Weinstein, B.M. Visualization and experimental analysis of blood vessel formation using transgenic zebrafish. Birth Defects Res. Part C Embryo Today Rev. 2007, 81, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Huisken, J.; Swoger, J.; Del Bene, F.; Wittbrodt, J.; Stelzer, E.H.K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004, 305, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, A.; Mickoleit, M.; Weber, M.; Huisken, J. Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development 2012, 139, 3242–3247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesage, D.; Angelini, E.D.; Bloch, I.; Funka-Lea, G. A review of 3D vessel lumen segmentation techniques: Models, features and extraction schemes. Med. Image Anal. 2009, 13, 819–845. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Ip, H.H.S.; Cheng, S.H.; Chan, P.K. A relational-tubular (ReTu) deformable model for vasculature quantification of zebrafish embryo from microangiography image series. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 2004, 28, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Cheng, S.H.; Chan, P.K.; Ip, H.H.S. Reconstruction and representation of caudal vasculature of zebrafish embryo from confocal scanning laser fluorescence microscopic images. Comput. Biol. Med. 2005, 35, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Ip, H.H.S. A statistical assembled deformable model (SAMTUS) for vasculature reconstruction. Comput. Biol. Med. 2009, 39, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Ip, H.; Feng, J.; Cheng, H. Automatic Segmentation and Tracking of Vasculature from Confocal Scanning Laser Fluorescence Microscope Images sequences using Multi-Orientation Dissection Sections. In Proceedings of the IEEE International Symposium on Biomedical Imaging, Washington, DC, USA, 7–10 July 2002; pp. 249–252. [Google Scholar]
- Schneider, S. Segmentation of Zebrafish Vasculature; Technical Report; MOSAIC Group: Towson, MD, USA, 2015. [Google Scholar]
- Tam, S.; Richmond, D.; Kaminker, J.; Modrusan, Z.; Martin-McNulty, B.; Cao, T.; Weimer, R.; Carano, R.D.; van Bruggen, N.; Watts, R. Death Receptors DR6 and TROY Regulate Brain Vascular Development. Dev. Cell 2012, 22, 403–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Jiang, L.; Li, C.; Hu, D.; Bu, J.W.; Cai, D.; Du, J.L. Haemodynamics-Driven Developmental Pruning of Brain Vasculature in Zebrafish. PLoS Biol. 2012, 10, e1001374. [Google Scholar] [CrossRef] [PubMed]
- Kugler, E.; Chico, T.; Armitage, P. Image Analysis in Light Sheet Fluorescence Microscopy Images of Transgenic Zebrafish Vascular Development. In Medical Image Understanding and Analysis. MIUA 2018; Communications in Computer and Information Science; Nixon, M., Mahmoodi, S., Zwiggelaar, R., Eds.; Springer: Cham, Switzerland, 2018; Volume 894, pp. 343–353. [Google Scholar]
- Chi, N.C.; Shaw, R.M.; De Val, S.; Kang, G.; Jan, L.Y.; Black, B.L.; Stainier, D.Y. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008, 22, 734–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, N.D.; Weinstein, B.M. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev. Biol. 2002, 248, 307–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Nakajima, S.; Atsumi, H.; Koller, T.; Gerig, G.; Yoshida, S.; Kikinis, R. 3D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. In CVRMed-MRCAS’97; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1997; pp. 213–222. [Google Scholar] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji—An Open Source platform for biological image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for Laboratory Use of Zebrafish (Brachydanio Rerio), 2nd ed.; University of Oregon Press: Eugene, OR, USA, 1993. [Google Scholar]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Lim, J. Two-Dimensional Signal and Image Processing; Prentice Hall: Englewood Cliffs, NJ, USA, 1990; pp. 469–476. [Google Scholar]
- Stelzer. Contrast, resolution, pixelation, dynamic range and signal-to-noise ratio: Fundamental limits to resolution in fluorescence light microscopy. J. Microsc. 1998, 189, 15–24. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Jain, A.K.; Dubes, R.C. Algorithms for Clustering Data; Prentice-Hall, Inc.: Upper Saddle River, NJ, USA, 1988. [Google Scholar]
- Arthur, D.; Vassilvitskii, S. K-means++: The Advantages of Careful Seeding. In Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms, SODA ’07, New Orleans, Louisiana, 7–9 January 2007; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2007; pp. 1027–1035. [Google Scholar]
- Nock, R.; Nielsen, F. Statistical region merging. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 1452–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, F.; Nock, R. On region merging: The statistical soundness of fast sorting, with applications. In Proceedings of the 2003 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Madison, WI, USA, 18–20 June 2003; Volume 2, pp. 19–26. [Google Scholar] [CrossRef]
- Sethian, J. Level Set Methods: Evolving Interfaces in Geometry, Fluid Mechanics, Computer Vision, and Materials Science; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- D’Agostino, R.B.; Belanger, A. A Suggestion for Using Powerful and Informative Tests of Normality. Am. Stat. 1990, 44, 316–321. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef] [Green Version]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kugler, E.; Plant, K.; Chico, T.; Armitage, P. Enhancement and Segmentation Workflow for the Developing Zebrafish Vasculature. J. Imaging 2019, 5, 14. https://doi.org/10.3390/jimaging5010014
Kugler E, Plant K, Chico T, Armitage P. Enhancement and Segmentation Workflow for the Developing Zebrafish Vasculature. Journal of Imaging. 2019; 5(1):14. https://doi.org/10.3390/jimaging5010014
Chicago/Turabian StyleKugler, Elisabeth, Karen Plant, Timothy Chico, and Paul Armitage. 2019. "Enhancement and Segmentation Workflow for the Developing Zebrafish Vasculature" Journal of Imaging 5, no. 1: 14. https://doi.org/10.3390/jimaging5010014
APA StyleKugler, E., Plant, K., Chico, T., & Armitage, P. (2019). Enhancement and Segmentation Workflow for the Developing Zebrafish Vasculature. Journal of Imaging, 5(1), 14. https://doi.org/10.3390/jimaging5010014