Molecularly Designed Ion-Imprinted Nanoparticles for Real-Time Sensing of Cu(II) Ions Using Quartz Crystal Microbalance
Abstract
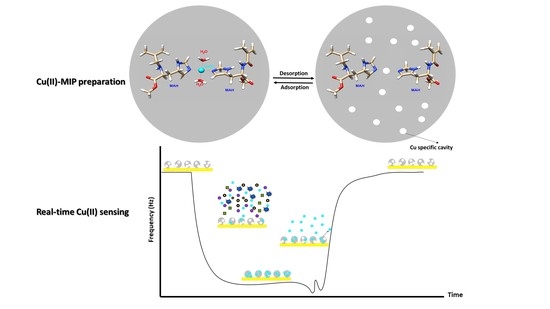
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Geometries of Cu(II)-MAH Complexes
2.3. Synthesis and Characterization of Cu(II)-MIP and NIP Nanoparticles
2.4. Conduction of Cu(II)-MIP and NIP Nanoparticles to QCM Nanosensor
2.5. MIP Efficiency of QCM Nanosensor
2.6. Equilibrium Isotherm Models, Detection, and Quantification Limits
2.7. Selectivity and Imprinting Efficiency of MIP-QCM Nanosensors
3. Results and Discussion
3.1. Characterization of Molecularly Designed Complexes and MIP Nanoparticles
3.2. Conduction of Cu(II)-MIP Nanoparticles to QCM Nanosensor
3.3. Efficiency of Cu(II)-MIP-Based QCM Nanosensor
3.4. Equilibrium Isotherm Models
3.5. Detection and Quantification Limits of Cu(II)-MIP QCM Nanosensor
3.6. Selectivity of Cu(II)-MIP QCM Nanosensors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. A Global Overview of National Regulations and Standards for Drinking-Water Quality, Verordnung Uber Die Qual. t von Wasser Für Den Menschl. Gebrauch (Trinkwasserverordnung—TrinkwV 2001); World Health Organization: Geneve, Switzerland, 2018. [Google Scholar]
- Choi, E.K.; Lim, D.G. Hepatic ischemia-reperfusion injury with respect to oxidative stress and inflammatory response: A narrative review. J. Yeungham Med. Sci. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Kehrer, J.P.; Klotz, L.O. Free radicals and related reactive species as mediators of tissue injury and disease: Implications for Health. Crit. Rev. Toxicol. 2015, 45, 765–798. [Google Scholar] [CrossRef] [PubMed]
- Bost, M.; Houdart, S.; Oberli, M.; Kalonji, E.; Huneau, J.F.; Margaritis, I. Dietary copper and human health: Current evidence and unresolved issues. J. Trace Elem. Med. Biol. 2016, 35, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, J.; Liu, X.; Gao, Y.; Ye, Z.; Li, G. Colorimetric copper(ii) ion sensor based on the conformational change of peptide immobilized onto the surface of gold nanoparticles. Anal. Methods 2014, 6, 2580–2585. [Google Scholar] [CrossRef]
- Baghban, N.; Yilmaz, E.; Soylak, M. Nanodiamond/MoS2 nanorod composite as a novel sorbent for fast and effective vortex-assisted micro solid phase extraction of lead(II) and copper(II) for their flame atomic absorption spectrometric detection. J. Mol. Liq. 2017, 234, 260–267. [Google Scholar] [CrossRef]
- Zheng, X.; Cheng, W.; Ji, C.; Zhang, J.; Yin, M. Detection of metal ions in biological systems: A review. Rev. Anal. Chem. 2020, 39, 231–246. [Google Scholar] [CrossRef]
- Lin, Y.S.; Lin, Y.F.; Nain, A.; Huang, Y.F.; Chang, H.T. A critical review of copper nanoclusters for monitoring of water quality. Sens. Actuators Rep. 2021, 3, 100026. [Google Scholar] [CrossRef]
- Saleem, M.; Rafiq, M.; Hanif, M.; Shaheen, M.A.; Seo, S.Y. A Brief Review on Fluorescent Copper Sensor Based on Conjugated Organic Dyes. J. Fluoresc. 2018, 28, 97–165. [Google Scholar] [CrossRef]
- Andaç, M.; Say, R.; Denizli, A. Molecular recognition based cadmium removal from human plasma. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2004, 811, 119–126. [Google Scholar] [CrossRef]
- Andaç, M.; Özyapi, E.; Şenel, S.; Say, R.; Denizli, A. Ion-selective imprinted beads for aluminum removal from aqueous solutions. Ind. Eng. Chem. Res. 2006, 45, 1780–1786. [Google Scholar] [CrossRef]
- Branger, C.; Meouche, W.; Margaillan, A. Recent advances on ion-imprinted polymers. React. Funct. Polym. 2013, 73, 859–875. [Google Scholar] [CrossRef]
- Spieker, E.; Lieberzeit, P.A. Molecular Imprinting Studies for Developing QCM-sensors for Bacillus Cereus. Procedia Eng. 2016, 168, 561–564. [Google Scholar] [CrossRef]
- Leibl, N.; Haubt, K.; Gonzato, C.; Duma, L. Molecularly Imprinted Polymers for Chemical Sensing: A Tutorial Review. Chemosensors 2022, 9, 123. [Google Scholar] [CrossRef]
- Ma, J.; Yan, M.; Feng, G.; Ying, Y.; Chen, G.; Shao, Y.; She, Y.; Wang, M.; Sun, J.; Zheng, L.; et al. An overview on molecular imprinted polymers combined with surface-enhanced Raman spectroscopy chemical sensors toward analytical applications. Talanta 2021, 225, 122031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Y.; Lu, X. Molecular imprinting technology for sensing foodborne pathogenic bacteria. Anal. Bioanal. Chem. 2021, 413, 4581–4598. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Ahmadi, Y.; Kim, K.H. Molecularly imprinted polymers for sensing gaseous volatile organic compounds: Opportunities and challenges. Environ. Pollut. 2022, 311, 119931. [Google Scholar] [CrossRef]
- Salha, D.; Andaç, M.; Denizli, A. Molecular docking of metal ion immobilized ligands to proteins in affinity chromatography. J. Mol. Recognit. 2021, 34, 1–11. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2009; pp. 1–20. [Google Scholar]
- Perdew, J.P. Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys. Rev. B 1986, 33, 8822. [Google Scholar] [CrossRef]
- Petersson, G.A.; Bennett, A.; Tensfeldt, T.G.; Al-Laham, M.A.; Shirley, W.A.; Mantzaris, J. A complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row elements. J. Chem. Phys. 1988, 89, 2193–2218. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Baydemir, G.; Derazshamshir, A.; Andaç, M.; Andaç, C.; Denizli, A. Reversible immobilization of glycoamylase by a variety of Cu 2+-chelated membranes. J. Appl. Polym. Sci. 2012, 126, 575–586. [Google Scholar] [CrossRef]
- Sener, G.; Ozgur, E.; Yilmaz, E.; Uzun, L.; Say, R.; Denizli, A. Quartz crystal microbalance based nanosensor for lysozyme detection with lysozyme imprinted nanoparticles. Biosens. Bioelectron. 2010, 26, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Babanejad, N.; Mfoafo, K.; Zhang, E.; Omidi, Y.; Razeghifard, R.; Omidian, H. Applications of cryostructures in the chromatographic separation of biomacromolecules. J. Chromatogr. A 2022, 1683, 463546. [Google Scholar] [CrossRef] [PubMed]
- Şenel, S.; Elmas, B.; Çamli, T.; Andaç, M.; Denizli, A. Poly(hydroxyethylmethacrylate-N-methacryloyl-(L)-histidine-methyl-ester) based metal-chelate affinity adsorbent for separation of lysozyme. Sep. Sci. Technol 2004, 39, 3783–3795. [Google Scholar] [CrossRef]
- Derazshamshir, A.; Baydemir, G.; Andac, M.; Say, R.; Galaev, I.Y.; Denizli, A. Molecularly imprinted PHEMA-based cryogel for depletion of hemoglobin from human blood. Macromol. Chem. Phys. 2010, 211, 657–668. [Google Scholar] [CrossRef]
- Demiralay, E.Ç.; Andaç, M.; Say, R.; Alsancak, G.; Denizli, A. Nickel(II)-Imprinted Monolithic Columns for Selective Nickel Recognition. J. Appl. Polym. Sci. 2010, 117, 3704–3714. [Google Scholar] [CrossRef]
- Tamahkar, E.; Bakhshpour, M.; Andaç, M.; Denizli, A. Ion imprinted cryogels for selective removal of Ni(II) ions from aqueous solutions. Sep. Purif. Technol. 2017, 179, 36–44. [Google Scholar] [CrossRef]
- Li, X.; Husson, S.M. Adsorption of dansylated amino acids on molecularly imprinted surfaces: A surface plasmon resonance study. Biosens. Bioelectron. 2006, 22, 336–348. [Google Scholar] [CrossRef]
- Abu-Alsoud, G.F.; Hawboldt, K.A.; Bottaro, C.S. Assessment of cross-reactivity in a tailor-made molecularly imprinted polymer for phenolic compounds using four adsorption isotherm models. J. Chromatogr. A. 2020, 1629, 461463. [Google Scholar] [CrossRef]
- Khayyun, T.S.; Mseer, A.H. Comparison of the experimental results with the Langmuir and Freundlich models for copper removal on limestone adsorbent. Appl. Water Sci. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Turiel, E.; Perez-Conde, C.; Martin-Esteban, A. Assessment of the cross-reactivity and binding sites characterisation of a propazine-imprinted polymer using the Langmuir-Freundlich isotherm. Analyst 2003, 128, 137–141. [Google Scholar] [CrossRef]
- Tan, S.S.; Kim, S.J.; Kool, E.T. Differentiating between fluorescence-quenching metal ions with polyfluorophore sensors built on a DNA backbone. J. Am. Chem. Soc. 2011, 133, 2664–2671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J.; Hwang, M.P.; Choi, M.; Seo, Y.; Jo, Y.; Son, J.; Hong, J.; Choi, J. Sensitive detection of copper ions via ion-responsive fluorescence quenching of engineered porous silicon nanoparticles. Sci. Rep. 2016, 6, 35565. [Google Scholar] [CrossRef] [PubMed]
- Awad, F.S.; AbouZied, K.M.; Bakry, A.M.; Abou El-Maaty, W.M.; El-Wakil, A.M.; El-Shall, M.S. Highly fluorescent hematoporphyrin modified graphene oxide for selective detection of copper ions in aqueous solutions. Anal. Chim. Acta 2020, 1140, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, C.; Wu, S.; Sun, C. Ion-imprinted polymer modified with carbon quantum dots as a highly sensitive copper(Ii) ion probe. Polymers 2021, 13, 1376. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Zhang, Y.; Xiang, Q.; Xu, J.; Li, H.; Xu, P.; Li, X. The real-time detection of trace-level Hg2+ in water by QCM loaded with thiol-functionalized SBA-15. Sens. Actuators B Chem. 2012, 166–167, 246–252. [Google Scholar] [CrossRef]









| Structures | Total Energy (Hartree) | Binding Energy (Hartree) |
|---|---|---|
| MAH-3-keto | −818.160 | |
| MAH-Cu-RingN-3aq-enol | −1243.188 | −0.6558 |
| MAH-Cu-RingN-5aq-enol | −1396.121 | −0.7499 |
| 2MAH-Cu-RingN-2aq-enol | −1984.963 | −0.7114 |
| 2MAH-Cu-RingN-4aq-enol | −2137.856 | −0.7655 |
| QCM Nanosensor Surface | Contact Angle (θ°) |
|---|---|
| Without Nanoparticles | 85.4 |
| Cu(II)-MIP1 | 41.8 |
| Cu(II)-MIP2 | 46.9 |
| Cu(II)-MIP1 QCM Nanosensor | |||||
| Langmuir | Freundlich | Langmuir–Freundlich | |||
| Δmmax | 0.287 | Δmmax | 0.165 | Δmmax | 0.292 |
| KD (µM) | 0.380 | 1/n | 0.179 | KD (µM) | 5.945 |
| KA (1/µM) | 2.630 | R2 | 0.845 | KA (1/µM) | 0.168 |
| R2 | 0.933 | R2 | 0.998 | ||
| Experimental Δmmax | 0.309 | ||||
| Cu(II)-MIP2 QCM nanosensor | |||||
| Langmuir | Freundlich | Langmuir–Freundlich | |||
| Δmmax | 0.606 | Δmmax | 0.226 | Δmmax | 1.071 |
| KD (µM) | 1.127 | 1/n | 0.266 | KD (µM) | 21.39 |
| KA (1/µM) | 0.887 | R2 | 0.802 | KA (1/µM) | 0.046 |
| R2 | 0.998 | R2 | 0.914 | ||
| Experimental Δmmax | 0.628 | ||||
| Cu(II)-MIP1 | Cu(II)-MIP2 | |
|---|---|---|
| Regression Equation | y = 0.0001x + 0.0743 | y = 0.0002x + 0.0914 |
| Standard Error of Slope | 2.09 × 10−7 | 3.95 × 10−7 |
| Standard Error of Intercept | 0.0008 | 0.0015 |
| Correlation Coefficient (r) | 0.999 | 0.999 |
| Calibration Range (nM) | 157.4–1574 | 157.4–1574 |
| Limit of Detection, LOD (nM) | 42.0 | 40.7 |
| Limit of Quantification, LOQ (nM) | 140.0 | 135.5 |
| Standard deviation | 0.0014 | 2.71 × 10−3 |
| Metal Ions | Cu(II)-MIP1 QCM Nanosensor | Cu(II)-MIP2 QCM Nanosensor | ||
|---|---|---|---|---|
| K Δm Cu(II)/Me(II) ** | K′ Δm MIP/NIP | K Δm Cu(II)/Me(II) | K′ Δm MIP/NIP | |
| Cu(II) | - | - | - | - |
| Co (II) | 8.1 | 17.1 | 6.9 | 29.5 |
| Fe (II) | 3.7 | 2.1 | 2.6 | 4.3 |
| Ni (II) | 7.0 | 4.7 | 5.8 | 11.5 |
| Zn (II) | 3.0 | 2.5 | 2.9 | 5.1 |
| Material | Sensor System | LOD | Sample | Ref |
|---|---|---|---|---|
| Porous silicon nanoparticles | Fluorescence quenching | 100 nM | Tap Water | [35] |
| Graphene oxide | Fluorescence sensing | 54 nM | Water | [36] |
| Ion-Imprinted polymer modified with carbon quantum dots | Fluorescence detection | 2.86 µM | Water | [37] |
| Thiol-functionalized SBA-15 mesoporous silica materials | QCM | N/A | Water | [38] |
| Molecularly designed nanoparticles | Gravimetric sensing | 42 nM | Aqueous media | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aydoğan, N.; Aylaz, G.; Bakhshpour, M.; Tugsuz, T.; Andaç, M. Molecularly Designed Ion-Imprinted Nanoparticles for Real-Time Sensing of Cu(II) Ions Using Quartz Crystal Microbalance. Biomimetics 2022, 7, 191. https://doi.org/10.3390/biomimetics7040191
Aydoğan N, Aylaz G, Bakhshpour M, Tugsuz T, Andaç M. Molecularly Designed Ion-Imprinted Nanoparticles for Real-Time Sensing of Cu(II) Ions Using Quartz Crystal Microbalance. Biomimetics. 2022; 7(4):191. https://doi.org/10.3390/biomimetics7040191
Chicago/Turabian StyleAydoğan, Nihan, Gülgün Aylaz, Monireh Bakhshpour, Tugba Tugsuz, and Müge Andaç. 2022. "Molecularly Designed Ion-Imprinted Nanoparticles for Real-Time Sensing of Cu(II) Ions Using Quartz Crystal Microbalance" Biomimetics 7, no. 4: 191. https://doi.org/10.3390/biomimetics7040191
APA StyleAydoğan, N., Aylaz, G., Bakhshpour, M., Tugsuz, T., & Andaç, M. (2022). Molecularly Designed Ion-Imprinted Nanoparticles for Real-Time Sensing of Cu(II) Ions Using Quartz Crystal Microbalance. Biomimetics, 7(4), 191. https://doi.org/10.3390/biomimetics7040191