Pilot Study: Safety and Performance Validation of an Ingestible Medical Device for Collecting Small Intestinal Liquid in Healthy Volunteers
Abstract
:1. Introduction
1.1. Rationale
1.2. Medical Device Description
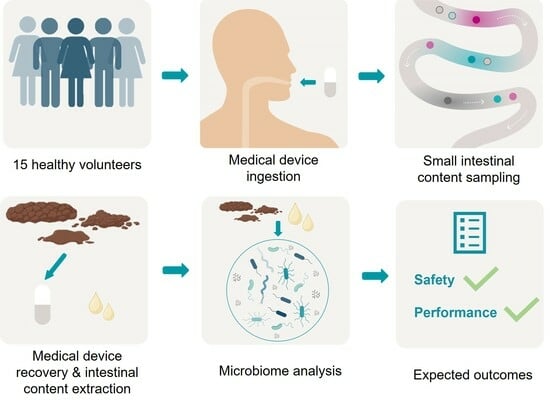
1.3. Aims and Objectives
1.4. Primary Outcomes
2. Experimental Design
2.1. Inclusion Criteria
2.2. Exclusion Criteria
2.3. Participants and Recruitment
2.4. Data Collection
2.5. Clinical Study Protocol
2.6. Multi-Omics Analysis
2.7. Sample Size
2.8. Statistical Analysis
2.9. Ethical Approval and Registration
3. Expected Results
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Postler, T.S.; Ghosh, S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. 2017, 26, 110–130. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Clooney, A.G.; Eckenberger, J.; Laserna-Mendieta, E.; Sexton, K.A.; Bernstein, M.T.; Vagianos, K.; Sargent, M.; Ryan, F.J.; Moran, C.; Sheehan, D.; et al. Ranking microbiome variance in inflammatory bowel disease: A large longitudinal intercontinental study. Gut 2021, 70, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Nhieu, J.T.V.; Furet, J.P. Microbial Dysbiosis in Colorectal Cancer (CRC) Patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef]
- Safari, Z.; Gérard, P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell. Mol. Life Sci. 2019, 76, 1541–1558. [Google Scholar] [CrossRef]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Araujo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of NAFLD is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatol. Baltim. Md. 2016, 63, 764. [Google Scholar] [CrossRef]
- Liu, J.; Tan, Y.; Cheng, H.; Zhang, D.; Feng, W.; Peng, C. Functions of Gut Microbiota Metabolites, Current Status and Future Perspectives. Aging Dis. 2022, 13, 1106–1126. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Tedelind, S.; Westberg, F.; Kjerrulf, M.; Vidal, A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: A study with relevance to inflammatory bowel disease. World J. Gastroenterol. 2007, 13, 2826–2832. [Google Scholar] [CrossRef] [PubMed]
- Delbaere, K.; Roegiers, I.; Bron, A.; Durif, C.; Van de Wiele, T.; Blanquet-Diot, S.; Marinelli, L. The small intestine: Dining table of host–microbiota meetings. FEMS Microbiol. Rev. 2023, 47, fuad022. [Google Scholar] [CrossRef] [PubMed]
- Young, W.; Maclean, P.; Dunstan, K.; Ryan, L.; Peters, J.; Armstrong, K.; Anderson, R.; Dewhurst, H.; van Gendt, M.; Dilger, R.N.; et al. Lacticaseibacillus rhamnosus HN001 alters the microbiota composition in the cecum but not the feces in a piglet model. Front. Nutr. 2022, 9, 1002369. [Google Scholar] [CrossRef] [PubMed]
- Sasso, J.M.; Ammar, R.M.; Tenchov, R.; Lemmel, S.; Kelber, O.; Grieswelle, M.; Zhou, Q.A. Gut Microbiome-Brain Alliance: A Landscape View into Mental and Gastrointestinal Health and Disorders. ACS Chem. Neurosci. 2023, 14, 1717–1763. [Google Scholar] [CrossRef]
- Villmones, H.C.; Haug, E.S.; Ulvestad, E.; Grude, N.; Stenstad, T.; Halland, A.; Kommedal, Ø. Species Level Description of the Human Ileal Bacterial Microbiota. Sci. Rep. 2018, 8, 4736. [Google Scholar] [CrossRef]
- Ahmed, S.; Macfarlane, G.T.; Fite, A.; McBain, A.J.; Gilbert, P.; Macfarlane, S. Mucosa-Associated Bacterial Diversity in Relation to Human Terminal Ileum and Colonic Biopsy Samples. Appl. Environ. Microbiol. 2007, 73, 7435–7442. [Google Scholar] [CrossRef]
- Villmones, H.C.; Svanevik, M.; Ulvestad, E.; Stenstad, T.; Anthonisen, I.L.; Nygaard, R.M.; Dyrhovden, R.; Kommedal, Ø. Investigating the human jejunal microbiota. Sci. Rep. 2022, 12, 1682. [Google Scholar] [CrossRef]
- Wang, M.; Ahrné, S.; Jeppsson, B.; Molin, G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 2005, 54, 219–231. [Google Scholar] [CrossRef] [PubMed]
- den Bogert, B.V.; Erkus, O.; Boekhorst, J.; de Goffau, M.; Smid, E.J.; Zoetendal, E.G.; Kleerebezem, M. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol. Ecol. 2013, 85, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Kastl, A.J.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Leite, G.; Morales, W.; Weitsman, S.; Celly, S.; Parodi, G.; Mathur, R.; Barlow, G.M.; Sedighi, R.; Millan, M.J.V.; Rezaie, A.; et al. The duodenal microbiome is altered in small intestinal bacterial overgrowth. PLoS ONE 2020, 15, e0234906. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Lennon, G.; Docherty, N.; Balfe, A.; Mulcahy, H.E.; Doherty, G.; O′Donoghue, D.; Hyland, J.M.; Shanahan, F.; Sheahan, K.; et al. Depth-Dependent Differences in Community Structure of the Human Colonic Microbiota in Health. PLoS ONE 2013, 8, e78835. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Lennon, G.; O’Sullivan, O.; Docherty, N.; Balfe, A.; Maguire, A.; Mulcahy, H.E.; Doherty, G.; O’Donoghue, D.; Hyland, J.; et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015, 64, 1553–1561. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef]
- Tang, Q.; Jin, G.; Wang, G.; Liu, T.; Liu, X.; Wang, B.; Cao, H. Current Sampling Methods for Gut Microbiota: A Call for More Precise Devices. Front. Cell. Infect. Microbiol. 2020, 10, 151. [Google Scholar] [CrossRef]
- Rehan, M.; Al-Bahadly, I.; Thomas, D.G.; Young, W.; Cheng, L.K.; Avci, E. Smart capsules for sensing and sampling the gut: Status, challenges and prospects. Gut 2023, 73, 186–202. [Google Scholar] [CrossRef]
- Fedoruk, M.J.; Guidotti, T.L. Gastrointestinal System. In Encyclopedia of Toxicology, 2nd ed.; Wexler, P., Ed.; Elsevier: New York, NY, USA, 2005; pp. 410–416. ISBN 978-0-12-369400-3. [Google Scholar]
- Jensen, B.A.H.; Heyndrickx, M.; Jonkers, D.; Mackie, A.; Millet, S.; Naghibi, M.; Pærregaard, S.I.; Pot, B.; Saulnier, D.; Sina, C.; et al. Small intestine vs. colon ecology and physiology: Why it matters in probiotic administration. CR Med. 2023, 4, 101190. [Google Scholar] [CrossRef]
- Steinberg, W.H.; Mina, F.A.; Pick, P.G.; Frey, G.H. Heidelberg Capsule I: In Vitro Evaluation of a New Instrument for Measuring Intragastric pH. J. Pharm. Sci. 1965, 54, 772–776. [Google Scholar] [CrossRef]
- Iddan, G.; Meron, G.; Glukhovsky, A.; Swain, P. Wireless capsule endoscopy. Nature 2000, 405, 417. [Google Scholar] [CrossRef] [PubMed]
- Pirkola, L.; Laatikainen, R.; Loponen, J.; Hongisto, S.-M.; Hillilä, M.; Nuora, A.; Yang, B.; Linderborg, K.M.; Freese, R. Low-FODMAP vs regular rye bread in irritable bowel syndrome: Randomized SmartPill® study. World J. Gastroenterol. 2018, 24, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.; Zhang, J.; Heimbach, T.; Penland, R.C.; Wanke, C.; Shimizu, J.; Kulmatycki, K. Novel Orally Swallowable IntelliCap® Device to Quantify Regional Drug Absorption in Human GI Tract Using Diltiazem as Model Drug. AAPS PharmSciTech 2014, 15, 1490–1497. [Google Scholar] [CrossRef]
- Vilz, T.O.; Pantelis, D.; Lingohr, P.; Fimmers, R.; Esmann, A.; Randau, T.; Kalff, J.C.; Coenen, M.; Wehner, S. SmartPill® as an objective parameter for determination of severity and duration of postoperative ileus: Study protocol of a prospective, two-arm, open-label trial (the PIDuSA study). BMJ Open 2016, 6, e011014. [Google Scholar] [CrossRef]
- Akpunonu, B.; Hummell, J.; Akpunonu, J.D.; Ud Din, S. Capsule endoscopy in gastrointestinal disease: Evaluation, diagnosis, and treatment. Clevel. Clin. J. Med. 2022, 89, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Valdastri, P.; Simi, M.; Webster, R.J. Advanced technologies for gastrointestinal endoscopy. Annu. Rev. Biomed. Eng. 2012, 14, 397–429. [Google Scholar] [CrossRef] [PubMed]
- Ruigrok, R.A.A.A.; Weersma, R.K.; Vich Vila, A. The emerging role of the small intestinal microbiota in human health and disease. Gut Microbes 2023, 15, 2201155. [Google Scholar] [CrossRef]
- Rump, A.; Kromrey, M.-L.; Scheuch, E.; Jannin, V.; Rehenbrock, L.; Tzvetkov, M.V.; Weitschies, W.; Grimm, M. In Vivo Evaluation of a Gastro-Resistant HPMC-Based “Next Generation Enteric” Capsule. Pharmaceutics 2022, 14, 1999. [Google Scholar] [CrossRef]
- Common Terminology Criteria for Adverse Events (CTCAE)|Protocol Development|CTEP. Available online: https://ctep.cancer.gov/protocolDevelopment/electronic_applications/ctc.htm (accessed on 25 September 2023).
- Mikolajczyk, A.E.; Watson, S.; Surma, B.L.; Rubin, D.T. Assessment of Tandem Measurements of pH and Total Gut Transit Time in Healthy Volunteers. Clin. Transl. Gastroenterol. 2015, 6, e100. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Shalon, D.; Culver, R.N.; Grembi, J.A.; Folz, J.; Treit, P.V.; Shi, H.; Rosenberger, F.A.; Dethlefsen, L.; Meng, X.; Yaffe, E.; et al. Profiling the human intestinal environment under physiological conditions. Nature 2023, 617, 581–591. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef]
- Menard, J.; Bagheri, S.; Menon, S.; Yu, Y.T.; Goodman, L.B. Noninvasive sampling of the small intestinal chyme for microbiome, metabolome and antimicrobial resistance genes in dogs, a proof of concept. Anim. Microbiome 2023, 5, 64. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tronel, A.; Silvent, A.-S.; Buelow, E.; Giai, J.; Leroy, C.; Proust, M.; Martin, D.; Le Gouellec, A.; Soranzo, T.; Mathieu, N. Pilot Study: Safety and Performance Validation of an Ingestible Medical Device for Collecting Small Intestinal Liquid in Healthy Volunteers. Methods Protoc. 2024, 7, 15. https://doi.org/10.3390/mps7010015
Tronel A, Silvent A-S, Buelow E, Giai J, Leroy C, Proust M, Martin D, Le Gouellec A, Soranzo T, Mathieu N. Pilot Study: Safety and Performance Validation of an Ingestible Medical Device for Collecting Small Intestinal Liquid in Healthy Volunteers. Methods and Protocols. 2024; 7(1):15. https://doi.org/10.3390/mps7010015
Chicago/Turabian StyleTronel, Alexandre, Anne-Sophie Silvent, Elena Buelow, Joris Giai, Corentin Leroy, Marion Proust, Donald Martin, Audrey Le Gouellec, Thomas Soranzo, and Nicolas Mathieu. 2024. "Pilot Study: Safety and Performance Validation of an Ingestible Medical Device for Collecting Small Intestinal Liquid in Healthy Volunteers" Methods and Protocols 7, no. 1: 15. https://doi.org/10.3390/mps7010015
APA StyleTronel, A., Silvent, A. -S., Buelow, E., Giai, J., Leroy, C., Proust, M., Martin, D., Le Gouellec, A., Soranzo, T., & Mathieu, N. (2024). Pilot Study: Safety and Performance Validation of an Ingestible Medical Device for Collecting Small Intestinal Liquid in Healthy Volunteers. Methods and Protocols, 7(1), 15. https://doi.org/10.3390/mps7010015