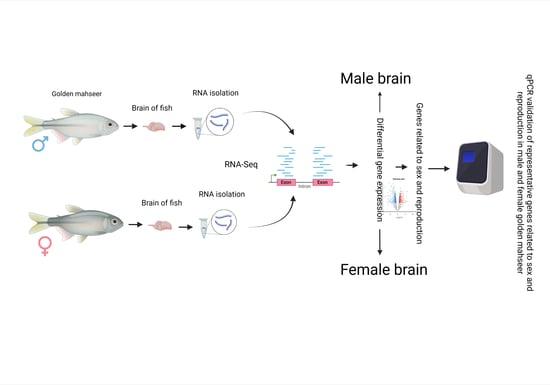
RNA-Seq Reveals Differential Gene Expression Patterns Related to Reproduction in the Golden Mahseer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Collection of Fish Sample
2.3. RNA Isolation, Library Preparation, and Sequencing
2.4. Transcriptome Assembly and Coding Sequence Prediction
2.5. Functional Annotation
2.6. Differential Expression of Genes
2.7. qPCR Validation of Differential Gene Expression
2.7.1. Fish Samples
2.7.2. RNA Isolation and Synthesis of cDNA
2.7.3. qPCR Assay
2.8. Relative Expression Estimation
2.9. Statistical Analysis
3. Results
3.1. Assembly and Functional Classification of Unigenes
3.2. Identification of Genes Showing Differential Sex Expression
3.3. Transcription Levels of Differential Gene Expressions Using qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Borella, M.I.; Chehade, C.; Costa, F.G.; de Jesus, L.W.O.; Cassel, M.; Batlouni, S.R. Chapter 14—The brain-pituitary-gonad axis and the gametogenesis. In Biology and Physiology of Freshwater Neotropical Fish; Baldisserotto, B., Urbinati, E.C., Cyrino, J.E.P., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 315–341. [Google Scholar] [CrossRef]
- Devlin, R.H.; Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 2002, 208, 191–364. [Google Scholar] [CrossRef]
- Bar, I.; Cummins, S.; Elizur, A. Transcriptome analysis reveals differentially expressed genes associated with germ cell and gonad development in the Southern bluefin tuna (Thunnus maccoyii). BMC Genom. 2016, 17, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zohar, Y.; Muñoz-Cueto, J.A.; Elizur, A.; Kah, O. Neuroendocrinology of reproduction in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 438–455. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, M.V.; Carrillo, M.; Felip, A. Expression of kisspeptins and their Receptors, gnrh-1/gnrhr-II-1a and gonadotropin genes in the brain of adult male and female European sea bass during different gonadal stages. Gen. Comp. Endocrinol. 2013, 187, 104–116. [Google Scholar] [CrossRef]
- Ando, H.; Sasaki, Y.; Okada, H.; Urano, A. Prepubertal increases in the levels of two salmon gonadotropin-releasing hormone mRNAs in the ventral telencephalon and preoptic area of masu salmon. Neurosci. Lett. 2001, 307, 93–96. [Google Scholar] [CrossRef]
- Mechaly, A.S.; Viñas, J.; Piferrer, F. Sex-specific changes in the expression of kisspeptin, kisspeptin receptor, gonadotropins and gonadotropin receptors in the Senegalese sole (Solea senegalensis) during a full reproductive cycle. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2012, 162, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Churcher, A.M.; Pujolar, J.M.; Milan, M.; Hubbard, P.C.; Martins, R.S.T.; Saraiva, J.L.; Huertas, M.; Bargelloni, L.; Patarnello, T.; Marino, I.A.M.; et al. Changes in the gene expression profiles of the brains of male European eels (Anguilla anguilla) during sexual maturation. BMC Genom. 2014, 15, 799. [Google Scholar] [CrossRef] [Green Version]
- Sahu, D.K.; Panda, S.P.; Meher, P.K.; Das, P.; Routray, P.; Sundaray, J.K.; Jayasankar, P.; Nandi, S. Construction, De-Novo Assembly and Analysis of Transcriptome for Identification of Reproduction-Related Genes and Pathways from Rohu, Labeo rohita (Hamilton). PLoS ONE 2015, 10, e0132450. [Google Scholar] [CrossRef] [Green Version]
- Shahi, N.; Singh, A.K.; Sahoo, M.; Mallik, S.K.; Thakuria, D. Molecular cloning, characterization and expression profile of kisspeptin1 and kisspeptin1 receptor at brain-pituitary-gonad (BPG) axis of golden mahseer, Tor putitora (Hamilton, 1822) during gonadal development. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 205, 13–29. [Google Scholar] [CrossRef]
- Pan, Z.; Zhu, C.; Chang, G.; Wu, N.; Ding, H.; Wang, H. Differential expression analysis and identification of sex-related genes by gonad transcriptome sequencing in estradiol-treated and non-treated Ussuri catfish Pseudobagrus ussuriensis. Fish Physiol. Biochem. 2021, 47, 565–581. [Google Scholar] [CrossRef]
- He, P.; Zhu, P.; Wei, P.; Zhuo, X.; Ma, Y.; Chen, X.; Lin, Y.; Xu, Y.; Luo, H.; Peng, J. Gonadal transcriptomic analysis and differentially expressed genes between the testes and ovaries in Trachinotus ovatus. Aquac. Fish. 2022, 7, 31–39. [Google Scholar] [CrossRef]
- Nazari, S.; Khoshkholgh, M.; Baeza, J.A. Comparative transcriptome sequencing analysis of the narrow-clawed crayfish Pontastacus leptodactylus (Eschscholtz, 1823) and discovery of candidate sex-related genes. Aquac. Rep. 2022, 25, 101235. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, X.; Zou, Y.; Yan, Z.; Huang, Y.; Zhu, Y.; Gao, J. De novo gonad transcriptome analysis of elongate loach (Leptobotia elongata) provides novel insights into sex-related genes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 42, 100962. [Google Scholar] [CrossRef]
- Sun, S.; Song, F.; Shi, L.; Zhang, K.; Gu, Y.; Sun, J.; Luo, J. Transcriptome analysis of differentially expressed circular RNAs in the testis and ovary of golden pompano (Trachinotus blochii). Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 45, 101052. [Google Scholar] [CrossRef]
- Pinder, A.C.; Britton, J.R.; Harrison, A.J.; Nautiyal, P.; Bower, S.D.; Cooke, S.J.; Lockett, S.; Everard, M.; Katwate, U.; Ranjeet, K.; et al. Mahseer (Tor spp.) fishes of the world: Status, challenges and opportunities for conservation. Rev. Fish Biol. Fish. 2019, 29, 417–452. [Google Scholar] [CrossRef] [Green Version]
- Barat, A.; Kumar, R.; Goel, C.; Singh, A.K.; Sahoo, P.K. De novo assembly and characterization of tissue-specific transcriptome in the endangered golden mahseer, Tor putitora. Meta Gene 2016, 7, 28–33. [Google Scholar] [CrossRef]
- Kumar, R.; Sahoo, P.K.; Barat, A. Transcriptome profiling and expression analysis of immune responsive genes in the liver of Golden mahseer (Tor putitora) challenged with Aeromonas hydrophila. Fish Shellfish Immunol. 2017, 67, 655–666. [Google Scholar] [CrossRef]
- Shahi, N.; Mallik, S.K.; Pande, J.; Das, P.; Singh, A.K. Spermatogenesis and related plasma androgen and progestin level in wild male golden mahseer, Tor putitora (Hamilton, 1822), during the spawning season. Fish Physiol. Biochem. 2015, 41, 909–920. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Development Core Team: Vienna, Austria, 2008. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Gong, Q.; Lai, J.; Song, M.; Du, J.; Deng, X. Gonadal transcriptome sequencing of the critically endangered Acipenser dabryanus to discover candidate sex-related genes. PeerJ 2018, 6, e5389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.-Y.; Gui, J.-F. Diverse and variable sex determination mechanisms in vertebrates. Sci. China Life Sci. 2018, 61, 1503–1514. [Google Scholar] [CrossRef]
- Mei, J.; Gui, J.-F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef] [Green Version]
- Tian, C.; Li, Z.; Dong, Z.; Huang, Y.; Du, T.; Chen, H.; Jiang, D.; Deng, S.; Zhang, Y.; Wanida, S.; et al. Transcriptome Analysis of Male and Female Mature Gonads of Silver Sillago (Sillago sihama). Genes 2019, 10, 129. [Google Scholar] [CrossRef] [Green Version]
- Mustapha, U.F.; Peng, Y.-X.; Huang, Y.-Q.; Assan, D.; Zhi, F.; Shi, G.; Huang, Y.; Li, G.-L.; Jiang, D.-N. Comparative transcriptome analysis of the differentiating gonads in Scatophagus argus. Front. Mar. Sci. 2022, 9, 962534. [Google Scholar] [CrossRef]
- Chen, W.; Liu, L.; Ge, W. Expression analysis of growth differentiation factor 9 (Gdf9/gdf9), anti-müllerian hormone (Amh/amh) and aromatase (Cyp19a1a/cyp19a1a) during gonadal differentiation of the zebrafish, Danio rerio†. Biol. Reprod. 2017, 96, 401–413. [Google Scholar] [CrossRef] [Green Version]
- Chi, W.; Gao, Y.; Hu, Q.; Guo, W.; Li, D. Genome-wide analysis of brain and gonad transcripts reveals changes of key sex reversal-related genes expression and signaling pathways in three stages of Monopterus albus. PLoS ONE 2017, 12, e0173974. [Google Scholar] [CrossRef] [Green Version]
- Cribbin, K.M.; Quackenbush, C.R.; Taylor, K.; Arias-Rodriguez, L.; Kelley, J.L. Sex-specific differences in transcriptome profiles of brain and muscle tissue of the tropical gar. BMC Genom. 2017, 18, 283. [Google Scholar] [CrossRef] [Green Version]
- Danzmann, R.G.; Kocmarek, A.L.; Norman, J.D.; Rexroad, C.E.; Palti, Y. Transcriptome profiling in fast versus slow-growing rainbow trout across seasonal gradients. BMC Genom. 2016, 17, 60. [Google Scholar] [CrossRef] [Green Version]
- Ellegren, H.; Parsch, J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007, 8, 689–698. [Google Scholar] [CrossRef]
- Yan, Y.-L.; Batzel, P.; Titus, T.; Sydes, J.; Desvignes, T.; BreMiller, R.; Draper, B.; Postlethwait, J.H. A Hormone That Lost Its Receptor: Anti-Müllerian Hormone (AMH) in Zebrafish Gonad Development and Sex Determination. Genetics 2019, 213, 529–553. [Google Scholar] [CrossRef]
- Xu, G.; Huang, T.; Jin, X.; Cui, C.; Li, D.; Sun, C.; Han, Y.; Mu, Z. Morphology, sex steroid level and gene expression analysis in gonadal sex reversal of triploid female (XXX) rainbow trout (Oncorhynchus mykiss). Fish Physiol. Biochem. 2016, 42, 193–202. [Google Scholar] [CrossRef]










| Primer Name | Primer Sequence (5′ to 3′) | Amplicon Length (bp) | PCR Efficiency (%) | Description |
|---|---|---|---|---|
| cyp19a1a | GCAGACGGTTCTCATACAGC TGTCTCTTCCAGCTTCTCCA | 154 | 96 | RNA-Seq validation and DEGs |
| amh | CCTGCAGACTCACAGAGTGG CTTGAGCAGCAAAACGGACC | 232 | 99 | RNA-Seq validation and DEGs |
| foxl3 | CACGTGGGCTCAAAATGTCC AGGAAAGCCTTGCGTCTGAA | 112 | 93 | Male and Female DEGs |
| dmrt2a | CCCCGGCAAAACTATGAGGT TTCCTGGAACGGATTCGGTG | 169 | 104 | RNA-Seq validation and DEGs |
| sox11b | ACCAAGATGACCAGTCACGGAA CGATTGTGGTGCAGGCGAG | 94 | 105 | Male and Female DEGs |
| gdf9 | CAGACCTGGAGGCCAGATTC TGCTTCTTCTCTGGTACGCC | 110 | 92 | Male and Female DEGs |
| sox9b | CTCCTGGAGAACACTCCGGT ATGAGTCTGCAGGCGTTGTG | 116 | 97 | RNA-Seq validation and DEGs |
| dax1 | TACGCGTACTTGAAAGGGGC TTAAGTGCCTGGTTCGCCTC | 104 | 97 | Male and Female DEGs |
| kif20 | AGCCAGCTCGAAAAACCTCT GGGTCCGTGCTGAGGATCTG | 154 | 97 | Male and Female DEGs |
| wt-1a | GGGGTGTGTAAGTCCTCTCTTC CGGATTTGGCGACCATCAAG | 138 | 99 | RNA-Seq validation and DEGs |
| aqp1 | CTCAGTGTTTGCCTGGGACA ACGAATCATTGCTGGTCCGA | 90 | 97 | RNA-Seq validation and DEGs |
| tkt | GACCACTACCACGAAGG AGGAACGTGGGACACAG | 128 | 98 | DEGs |
| sox9a | AATCTGAAGACGGCAGCGAA ATCGAATCGAATGGCGAGTCA | 156 | 99 | DEGs |
| klhl6 | GGACCAAAGTTCGTGGAGGT AGCCACGTAAAGGAAACCGT | 188 | 103 | Male and Female DEGs |
| star | GTGGAACCCCAATGTCAAACA AAGAACCTGAGAGGGACCAAA | 232 | 99 | RNA-Seq validation and DEGs |
| cxcl2 | CACATCAGCGGAGGACACAT GGGTGTAACTCCGTAGAGCG | 147 | 99 | Male and Female DEGs |
| ß-actin | TGTCCCTTCCCTTATGGCCT CATCCCAGTCCCTAAAGTGCT | 72 | 100 | Reference gene |
| hsd11b3 | GCATATGCGTCGCGTTCATT AATGGTACGCCAACTGCTCA | 119 | 99 | RNA-Seq validation |
| nr3c2 | TGTTTTGTGGCTTAGTAAATG GAGTTCCCTGGGTGATTGGG | 112 | 98 | RNA-Seq validation |
| esr2a | CAGCTCCCGTTTGTCTCACT GTTTAGGGTCCGTGCTGTGA | 105 | 100 | RNA-Seq validation |
| gpcr | TACGGTGTTTGGGTGTTGCT CCCCTTGCTGTGGAAGTGAA | 105 | 97 | RNA-Seq validation |
| mab3 | CTGATGGAGCTCAAGACGCA GTCGACAGAGAAGGTTCCCG | 102 | 98 | RNA-Seq validation |
| foxf2 | ACCGCATCTGACTTCCGTTT TTTTGGAGAGCCGAGTGCAT | 152 | 100 | RNA-Seq validation |
| hsd17b2 | CCTGCTTTATTTGTGAGCCA AGCACAAAGGCCTGAGGTGA | 112 | 99 | RNA-Seq validation |
| fgf13 | GAGAAATCAAACGCCTGCCG TGGAGCCGAAAAGCTTGACT | 109 | 100 | RNA-Seq validation |
| tdrd3 | CTACGAAGAACCTCCCCACG GGACTTTGCCTCTTCACCGA | 93 | 96 | RNA-Seq validation |
| tacc2 | TTTGTGAAGATGCTGGCGTTG GTCCGAAAGGCTCGTCTCTT | 102 | 99 | RNA-Seq validation |
| spag16 | GTTGGGCATGGGTTTGACG TTTGGAAGGCCCAACACCTT | 192 | 97 | RNA-Seq validation |
| ruvbl2 | AGGTGGCAACCACAAAGGTT GGCTCCAGAGCATCATCCAA | 106 | 103 | RNA-Seq validation |
| nup98 | CACCTACCGCTCAACCAACA CCCCCATCGAAGCATTTTGC | 123 | 96 | RNA-Seq validation |
| tbrg1 | CTCGCAGAGGCAAACCCTTA CAGGGCAGCTCTGGATTAGG | 99 | 92 | RNA-Seq validation |
| ddx19 | AGCTGCAGCGGAATCGATAA TTGCTTCGGTCTTCGCTTCT | 101 | 102 | RNA-Seq validation |
| sox10 | GCTCAACTGCTACGACTGGA AAGACCAGGTGAAAGACGTGA | 187 | 100 | RNA-Seq validation |
| iqce | TACATCCGTACAGCGACGAC CCGTTTAGAAAAGCAGCGGG | 111 | 98 | RNA-Seq validation |
| Organ | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|---|
| Male brain | 20,610,105 | 3,762,376,800 | 19,678,723 | 3,367,741,635 | 0.0257 | 98.05 | 94.29 | 50.08 |
| Female brain | 21,541,256 | 5,674,182,705 | 20,456,857 | 5,237,821,465 | 0.0248 | 98.12 | 93.58 | 49.91 |
| Description | Male Brain | Female Brain |
|---|---|---|
| Number of reads | 20,610,105 | 21,541,256 |
| Total number of bases | 3,762,376,800 | 5,674,182,705 |
| Number of transcripts contigs | 39,047 | 75,736 |
| Total transcript contigs length (bp) | 58,794,838 | 311,938,504 |
| Average transcript contigs length (bp) | 1505 | 1742 |
| Maximum length of transcript contigs | 14,972 | 18,509 |
| N50 value | 1646 | 2729 |
| Number of unigenes | 26,989 | 55,600 |
| Maximum length of unigenes | 11,289 | 18,509 |
| Total unigenes length (bp) | 26,397,813 | 87,317,771 |
| Average unigenes length (bp) | 978 | 1570 |
| Gene Symbol | Gene Description | p-Value | Log2FC Female/Male |
|---|---|---|---|
| Sex determination, gonadal development, and neuronal development | |||
| sox10 | SRY-box transcription factor 10 | 2.63 × 10−18 | 0.67 |
| sox19a | SRY-box transcription factor 19a | 3.01 × 10−11 | 1.15 |
| sox4 | SRY-box transcription factor 4 | 1.15 × 10−29 | 0.72 |
| sox11a | SRY-box transcription factor 11a | 4.06 × 10−22 | 0.91 |
| sox21b | SRY-box transcription factor 21b | 3.02 × 10−6 | 1.23 |
| sox4b | SRY-box transcription factor 4b | 6.71 × 10−16 | 0.76 |
| sox5x3 | SRY-box transcription factor 5 isoform x3 | 2.31 × 10−9 | 1.92 |
| sox7 | SRY-box transcription factor 7 | 1.42 × 10−10 | 1.01 |
| sox3 | SRY-box transcription factor 3 | 1.67 × 10−8 | 0.67 |
| wnt5 | Wingless-type MMTV integration site family, member 5 | 3.12 × 10−6 | 11.78 |
| dmrt2a | Double sex and mab-3-related transcription factor 2a | 3.89 × 10−7 | −17.93 |
| dmrta2 | Double sex and mab-3-related transcription factor a2 | 7.55 × 10−17 | −19.12 |
| dmrta1 | Double sex and mab-3-related transcription factor a1 | 6.35 × 10−12 | 1.56 |
| dmrt2b | Double sex and mab-3-related transcription factor 2b | 3.65 × 10−19 | −17.04 |
| foxl1 | Forkhead box protein l1 | 1.23 × 10−4 | 1.67 |
| foxf2 | Forkhead box protein f2 | 8.81 × 10−15 | 3.90 |
| foxn2 | Forkhead box protein n2 | 1.91 × 10−27 | −8.78 |
| foxj3 | Forkhead box protein J3 | 5.03 × 10−22 | 7.67 |
| foxd1 | Forkhead box protein D1 | 3.76 × 10−22 | 4.78 |
| foxn2 | Forkhead box protein N2 | 1.63 × 10−20 | 2.99 |
| fgf11 | Fibroblast growth factor 11 | 5.09 × 10−22 | −6.76 |
| fgf8b | Fibroblast growth factor 8b | 2.83 × 10−34 | 3.24 |
| wt-1a | Wilms tumor protein 1a | 3.99 × 10−16 | 5.03 |
| sry | Sex determining region Y | 3.98 × 10−43 | −9.85 |
| mab3 | Mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase | 1.86 × 10−34 | 6.39 |
| alcam | Activated leukocyte cell adhesion molecule | 2.76 × 10−9 | 1.93 |
| hint2 | Histidine triad nucleotide-binding protein 2 | 2.42 × 10−14 | −11.26 |
| hint3 | Histidine triad nucleotide-binding protein 3 | 2.55 × 10−57 | −11.39 |
| xicof20 | Oocyte zinc finger protein 20 | 6.35 × 10−100 | 19.94 |
| xicof6 | Oocyte zinc finger protein 6 | 8.02 × 10−13 | 15.45 |
| xicof26 | Oocyte zinc finger protein 26 | 7.66 × 10−148 | 13.45 |
| xicgf | Gastrula zinc finger protein | 3.09 × 10−5 | −0.75 |
| tdrd | Todor and KH domain-containing protein | 1.51 × 10−14 | 6.55 |
| spag16 | Sperm associated antigen 16 protein | 3.45 × 10−47 | 0.13 |
| spag1 | Sperm associated antigen 1 | 5.03 × 10−20 | 0.03 |
| spag2 | Sperm specific antigen 2 | 3.46 × 10−13 | 0.12 |
| sat1 | Spermidine/spermine N1-acetyltransferase | 8.09 × 10−23 | −15.78 |
| spef2 | Sperm flagellar protein 2 | 5.09 × 10−43 | −12.38 |
| ef1a2 | Elongation factor 1-alpha2 | 1.11 × 10−18 | 1.88 |
| cxcl2 | C-X-C motif chemokine ligand 2 | 3.74 × 10−26 | 2.96 |
| rgs4 | Regulator of G protein signaling 4 | 3.10 × 10−12 | 1.40 |
| muc13 | Mucin 13 | 1.87 × 10−17 | −6.89 |
| trim39 | Tripartite motif containing 39 | 1.24 × 10−5 | 2.91 |
| tacc2 | Transforming acidic coiled-coil-containing protein 2 | 5.75 × 10−17 | 3.15 |
| klhl6 | Kelch-like protein 6 | 3.67 × 10−19 | 1.37 |
| klhl4 | Kelch-like protein 4 | 6.31 × 10−2 | 0.45 |
| bcl-2 | Apoptosis regulator factor 2 | 9.05 × 10−22 | 7.45 |
| bcl-2a | Bcl2 related ovarian killer protein homolog a isoformx2 | 1.88 × 10−41 | −6.23 |
| phb2 | Prohibitin-2 | 2.31 × 10−6 | 0.67 |
| map7 | Ensconsin | 6.53 × 10−13 | 4.70 |
| aqp1 | Aquaporin 1 | 1.21 × 10−10 | 15.06 |
| tgfb1 | Transforming growth factor beta 1 | 2.43 × 10−8 | 1.63 |
| rdh10 | Retinol dehydrogenase 10 | 3.01 × 10−5 | 3.16 |
| nipbl | Nipped-b-like protein | 3.12 × 10−6 | 5.23 |
| Hormone receptors and steroidogenesis | |||
| hsd17b2 | Hydroxysteroid 17-beta dehydrogenase 2 | 1.23 × 10−11 | 2.33 |
| hsd17b1 | Hydroxysteroid 17-beta dehydrogenase 1 | 3.65 × 10−9 | −1.93 |
| hsd20b | Hydroxysteroid 20-beta dehydrogenase | 5.07 × 10−11 | 6.09 |
| hsd3b7 | Hydroxysteroid 3-beta dehydrogenase 7 | 3.76 × 10−26 | 2.87 |
| hsd17b14 | Hydroxysteroid 17-beta dehydrogenase 14 | 2.16 × 10−9 | 2.89 |
| hsd17b4 | Hydroxysteroid 17-beta dehydrogenase 4 | 5.22 × 10−13 | 4.01 |
| hsd17b12 | Hydroxysteroid 17-beta-dehydrogenase 12 | 1.19 × 10−14 | 7.61 |
| hsd17b2 | Hydroxysteroid 17-beta-dehydrogenase 2 | 2.76 × 10−6 | −1.67 |
| esrα | Estrogen receptor alpha | 1.99 × 10−21 | 4.54 |
| esr1 | Estrogen receptor 1 | 3.41 × 10−77 | 5.90 |
| esr2a | Estrogen receptor 2a | 6.61 × 10−11 | 13.98 |
| esr2b | Estrogen receptor 2b | 2.67 × 10−17 | 9.45 |
| cyp19a1a | Cytochrome P450, family 19, subfamily A, polypeptide 1a | 5.51 × 10−12 | 15.56 |
| cyp20a1a | Cytochrome P450 family 20, subfamily A, polypeptide 1a | 6.31 × 10−19 | 12.78 |
| nr3c1 | Glucocorticoid receptor 1 | 1.99 × 10−9 | 5.13 |
| nr3c2 | Mineralocorticoid receptor 2 | 2.78 × 10−19 | 3.25 |
| tkt | Transketolase | 1.66 × 10−7 | 3.61 |
| gpcr | G-protein coupled estrogen receptor 1 | 9.23 × 10−11 | 2.37 |
| star | Steroidogenic acute regulatory protein | 5.87 × 10−8 | 6.91 |
| cyp11a1 | Cytochrome P450, family 11, subfamily A, polypeptide 1 | 2.99 × 10−21 | 4.83 |
| cyp11c1 | Cytochrome P450, family 11, subfamily C, polypeptide 1 | 3.06 × 10−1 | 5.75 |
| cyp11b2 | Cytochrome P450, family 11, subfamily B, polypeptide 1 | 7.61 × 10−22 | 4.34 |
| npb | Neuropeptide B receptor | 1.23 × 10−12 | −3.56 |
| npy | Neuropeptide Y receptor | 7.43 × 10−26 | −11.09 |
| hgfr | Hepatocyte growth factor receptor 1 | 5.12 × 10−1 | 2.90 |
| fgf | Fibroblast growth factor receptor | 2.99 × 10−34 | 3.45 |
| tbrg | Transforming growth factor-beta receptor | 1.23 × 10−72 | 13.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahi, N.; Singh, B.; Mallik, S.K.; Sarma, D.; Surachetpong, W. RNA-Seq Reveals Differential Gene Expression Patterns Related to Reproduction in the Golden Mahseer. Fishes 2023, 8, 352. https://doi.org/10.3390/fishes8070352
Shahi N, Singh B, Mallik SK, Sarma D, Surachetpong W. RNA-Seq Reveals Differential Gene Expression Patterns Related to Reproduction in the Golden Mahseer. Fishes. 2023; 8(7):352. https://doi.org/10.3390/fishes8070352
Chicago/Turabian StyleShahi, Neetu, Bhupendra Singh, Sumanta Kumar Mallik, Debajit Sarma, and Win Surachetpong. 2023. "RNA-Seq Reveals Differential Gene Expression Patterns Related to Reproduction in the Golden Mahseer" Fishes 8, no. 7: 352. https://doi.org/10.3390/fishes8070352
APA StyleShahi, N., Singh, B., Mallik, S. K., Sarma, D., & Surachetpong, W. (2023). RNA-Seq Reveals Differential Gene Expression Patterns Related to Reproduction in the Golden Mahseer. Fishes, 8(7), 352. https://doi.org/10.3390/fishes8070352