Transcriptome-Based Analysis of the Liver Response Mechanism of Black Porgy (Acanthopagrus schlegelii) to Stocking Density
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals
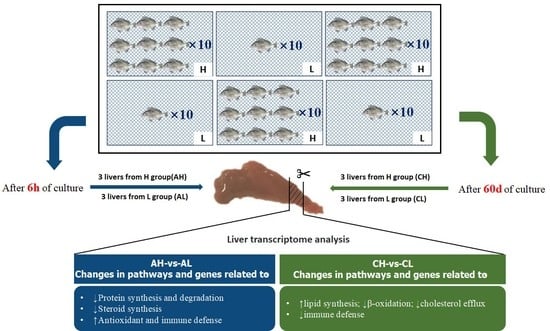
2.3. Experimental Design and Sample Collection
2.4. Total RNA Extraction, cDNA Library Construction, and Sequencing
2.5. Sequencing Data Processing and Analysis
2.6. RT-qPCR Verification
3. Results
3.1. Transcriptome Sequencing Data Quality Analysis
3.2. Analysis of DEGs between Groups
3.3. GO Enrichment Analysis of DEGs
3.4. KEGG Enrichment Analysis of DEGs
3.5. Validation of RNA-Seq Results
4. Discussion
4.1. Effects of High Stocking Density on Protein Metabolism-Related Pathways
4.2. Effects of High Stocking Density on Lipid Metabolism-Related Pathways
4.3. Effects of High Stocking Density on Immunity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2022; FAO: Rome, Italy, 2022; ISBN 978-92-5-136364-5. [Google Scholar]
- Garcia, F.; Romera, D.M.; Gozi, K.S.; Onaka, E.M.; Fonseca, F.S.; Schalch, S.H.C.; Candeira, P.G.; Guerra, L.O.M.; Carmo, F.J.; Carneiro, D.J.; et al. Stocking Density of Nile Tilapia in Cages Placed in a Hydroelectric Reservoir. Aquaculture 2013, 410–411, 51–56. [Google Scholar] [CrossRef]
- Mishal, P.; Karnatak, G.; Das, B.K.; Tayung, T.; Kumari, S.; Sarkar, U.K.; Das, A.K.; Ali, Y. Impact of Stocking Density on Growth, Feed Utilization and Survival of Cage Reared Minor Carp, Labeo bata (Hamilton, 1822) in Maithon Reservoir, India. Aquaculture 2021, 532, 736078. [Google Scholar] [CrossRef]
- Zhao, H.H.; Soufan, O.; Xia, J.G.; Tang, R.; Li, L.; Li, D.P. Transcriptome and Physiological Analysis Reveal Alterations in Muscle Metabolisms and Immune Responses of Grass Carp (Ctenopharyngodon idellus) Cultured at Different Stocking Densities. Aquaculture 2019, 503, 186–197. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Ni, J.J.; Nie, Z.J.; Gao, J.C.; Sun, Y.; Shao, N.L.; Li, Q.J.; Hu, J.W.; Xu, P.; Xu, G.C. Effects of Stocking Density on Growth, Serum Parameters, Antioxidant Status, Liver and Intestine Histology and Gene Expression of Largemouth Bass (Micropterus salmoides) Farmed in the in-Pond Raceway System. Aquac. Res. 2020, 51, 5228–5240. [Google Scholar] [CrossRef]
- Liu, Q.; Hou, Z.S.; Wen, H.S.; Li, J.F.; He, F.; Wang, J.H.; Guan, B.; Wang, Q.L. Effect of Stocking Density on Water Quality and (Growth, Body Composition and Plasma Cortisol Content) Performance of Pen-Reared Rainbow Trout (Oncorhynchus mykiss). J. Ocean. Univ. China 2016, 15, 667–675. [Google Scholar] [CrossRef]
- Zhou, F.; Song, W.X.; Shao, Q.J.; Peng, X.; Xiao, J.X.; Hua, Y.; Owari, B.N.; Zhang, T.Z.; Ng, W.K. Partial Replacement of Fish Meal by Fermented Soybean Meal in Diets for Black Sea Bream, Acanthopagrus schlegelii, Juveniles. J. World Aquac. Soc. 2011, 42, 184–197. [Google Scholar] [CrossRef]
- Jin, M.; Pan, T.T.; Cheng, X.; Zhu, T.T.; Sun, P.; Zhou, F.; Ding, X.Y.; Zhou, Q.C. Effects of Supplemental Dietary L-Carnitine and Bile Acids on Growth Performance, Antioxidant and Immune Ability, Histopathological Changes and Inflammatory Response in Juvenile Black Seabream (Acanthopagrus schlegelii) Fed High-Fat Diet. Aquaculture 2019, 504, 199–209. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Zhang, K.; Chen, S.; Zhang, Z.W.; Zhang, J.Y.; You, X.X.; Bian, C.; Xu, J.; Jia, C.F.; Qiang, J.; et al. Draft Genome of the Protandrous Chinese Black Porgy, Acanthopagrus Schlegelii. Gigascience 2018, 7, giy012. [Google Scholar] [CrossRef] [Green Version]
- Jia, C.F.; Zhu, F.; Qiu, M.; Sun, R.J.; Liu, H.L.; Zhang, Z.W.; Chen, S.Y.; Xu, J.; Zhang, Z.Y. Effects of low-salinity water cultivation on nutrient components of black porgy (Acanthopagrus schlegelii). Sci. Technol. Food Ind. 2020, 41, 284—288, 294. [Google Scholar] [CrossRef]
- Garcia Garcia, B.; Basurco, B.; Lovatelli, A. Current Status of Sparidae Aquaculture. In Sparidae: Biology and Aquaculture of Gilthead Sea Bream and Other Species; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; pp. 1–50. ISBN 978-1-4051-9772-4. [Google Scholar]
- Qian, X.; Ba, Y.; Zhuang, Q.F.; Zhong, G.F. RNA-Seq Technology and Its Application in Fish Transcriptomics. OMICS A J. Integr. Biol. 2014, 18, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, J. Signal Transduction Pathways and the Control of Cellular Responses to External Stimuli. In Functional Metabolism: Regulation and Adaptation, 1st ed.; Storey, B., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005; pp. 87–123. ISBN 978-0-471-67558-7. [Google Scholar]
- De Nadal, E.; Ammerer, G.; Posas, F. Controlling Gene Expression in Response to Stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef]
- Rodriguez Barreto, D.; Rey, O.; Uren-Webster, T.M.; Castaldo, G.; Consuegra, S.; Garcia de Leaniz, C. Transcriptomic Response to Aquaculture Intensification in Nile Tilapia. Evol. Appl. 2019, 12, 1757–1771. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Bao, P.B.; Tang, B.J. Transcriptome Analysis and Discovery of Genes Involved in Immune Pathways in Large Yellow Croaker (Larimichthys crocea) under High Stocking Density Stress. Fish Shellfish. Immunol. 2017, 68, 332–340. [Google Scholar] [CrossRef]
- He, Y.; Yu, H.Y.; Zhao, H.G.; Zhu, H.; Zhang, Q.J.; Wang, A.Q.; Shen, Y.B.; Xu, X.Y.; Li, J.L. Transcriptomic Analysis to Elucidate the Effects of High Stocking Density on Grass Carp (Ctenopharyngodon idella). BMC Genom. 2021, 22, 620. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.X.; Gao, G.T.; Palti, Y.; Cleveland, B.M.; Weber, G.M.; Iii, C.E.R. RNA-Seq Analysis of Early Hepatic Response to Handling and Confinement Stress in Rainbow Trout. PLoS ONE 2014, 9, e88492. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Muros, M.J.; Sánchez, B.; Barroso, F.G.; Toniolo, M.; Trenzado, C.E.; Sanz Rus, A. Effects of Rearing Conditions on Behavioural Responses, Social Kinetics and Physiological Parameters in Gilthead Sea Bream Sparus aurata. Appl. Anim. Behav. Sci. 2017, 197, 120–128. [Google Scholar] [CrossRef]
- Fanouraki, E.; Divanach, P.; Pavlidis, M. Baseline Values for Acute and Chronic Stress Indicators in Sexually Immature Red Porgy (Pagrus pagrus). Aquaculture 2007, 265, 294–304. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Meth. Mol. Biol. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wei, M.L.; Shen, M.J.; Wang, Y.; Gao, B.; Sun, R.J.; Qin, Y.L.; Jiang, J.B.; Zhou, J.L.; Shen, J.; Tang, X.J.; et al. A Cold-Inducible RNA Binding Protein Gene from Acanthopagrus schlegelii: Molecular Characterization, Expression, and Association with Apoptosis to Low-Temperature Stress. Aquac. Res. 2023, 2023, e1697400. [Google Scholar] [CrossRef]
- Li, X.J.; Shen, Y.D.; Bao, Y.G.; Wu, Z.X.; Yang, B.Q.; Jiao, L.F.; Zhang, C.D.; Tocher, D.R.; Zhou, Q.C.; Jin, M. Physiological Responses and Adaptive Strategies to Acute Low-Salinity Environmental Stress of the Euryhaline Marine Fish Black Seabream (Acanthopagrus schlegelii). Aquaculture 2022, 554, 738117. [Google Scholar] [CrossRef]
- Shi, Q.C.; Yu, C.Q.; Zhu, D.S.; Li, S.K.; Wen, X.B. Effects of Dietary Sargassum Horneri on Resisting Hypoxia Stress, Which Changes Blood Biochemistry, Antioxidant Status, and Hepatic HSP mRNA Expressions of Juvenile Black Sea Bream Acanthopagrus schlegelii. J. Appl. Phycol. 2020, 32, 3457–3466. [Google Scholar] [CrossRef]
- Tegomo, F.A.; Zhong, Z.W.; Njomoue, A.P.; Okon, S.U.; Ullah, S.; Gray, N.A.; Chen, K.; Sun, Y.X.; Xiao, J.X.; Wang, L.; et al. Experimental Studies on the Impact of the Projected Ocean Acidification on Fish Survival, Health, Growth, and Meat Quality; Black Sea Bream (Acanthopagrus schlegelii), Physiological and Histological Studies. Animals 2021, 11, 3119. [Google Scholar] [CrossRef]
- Qiu, Y.Y.; Zhang, Z.Y.; Chen, S.Y.; Ni, K.W.; Jia, C.F.; Meng, Q.; Zhu, F.; Zhang, Z.W.; Tang, X.J. Comparative Study on feeding frequency of hybrid F2 of Acanthopagrus schlegelii ♀ × Pagrus major♂ and A. schlegelii. South China Fish. Sci. 2022, 18, 59–67. [Google Scholar] [CrossRef]
- Portz, D.E.; Woodley, C.M.; Cech, J.J. Stress-Associated Impacts of Short-Term Holding on Fishes. Rev. Fish Biol. Fish. 2006, 16, 125–170. [Google Scholar] [CrossRef]
- Qi, L.J.; Chen, Y.D.; Shi, K.P.; Ma, H.; Wei, S.; Sha, Z.X. Combining of Transcriptomic and Proteomic Data to Mine Immune-Related Genes and Proteins in the Liver of Cynoglossus semilaevis Challenged with Vibrio anguillarum. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 39, 100864. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Xu, G.C.; Tang, Y.K.; Su, S.Y.; Wang, Y.P.; Zhu, Z.X. Investigation of the Molecular Mechanisms of Antioxidant Damage and Immune Response Downregulation in Liver of Coilia nasus under Starvation Stress. Front. Endocrinol. 2021, 12, 622315. [Google Scholar] [CrossRef] [PubMed]
- Ghafoory, S.; Breitkopf-Heinlein, K.; Li, Q.; Scholl, C.; Dooley, S.; Wölfl, S. Zonation of Nitrogen and Glucose Metabolism Gene Expression upon Acute Liver Damage in Mouse. PLoS ONE 2013, 8, e78262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.K.; Wang, X.L.; Sun, F.Y.; Zhang, J.R.; Feng, J.B.; Liu, H.; Rajendran, K.V.; Sun, L.Y.; Zhang, Y.; Jiang, Y.L.; et al. RNA-Seq Reveals Expression Signatures of Genes Involved in Oxygen Transport, Protein Synthesis, Folding, and Degradation in Response to Heat Stress in Catfish. Physiol. Genom. 2013, 45, 462–476. [Google Scholar] [CrossRef] [Green Version]
- Tian, C.X.; Lin, X.H.; Saetan, W.; Huang, Y.; Shi, H.J.; Jiang, D.N.; Chen, H.P.; Deng, S.P.; Wu, T.L.; Zhang, Y.L.; et al. Transcriptome Analysis of Liver Provides Insight into Metabolic and Translation Changes under Hypoxia and Reoxygenation Stress in Silver Sillago (Sillago sihama). Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 36, 100715. [Google Scholar] [CrossRef]
- Evans, T.G.; Kültz, D. The Cellular Stress Response in Fish Exposed to Salinity Fluctuations. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2020, 333, 421–435. [Google Scholar] [CrossRef]
- Naderi, M.; Keyvanshokooh, S.; Ghaedi, A.; Salati, A.P. Effect of Acute Crowding Stress on Rainbow Trout (Oncorhynchus mykiss): A Proteomics Study. Aquaculture 2018, 495, 106–114. [Google Scholar] [CrossRef]
- McClain, W.H. Rules That Govern tRNA Identity in Protein Synthesis. J. Mol. Biol. 1993, 234, 257–280. [Google Scholar] [CrossRef]
- Fromont Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome Assembly in Eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.L.; Zou, Z.Y.; Li, D.Y.; Xiao, W.; Yu, J.; Chen, B.L.; Xue, L.Y.; Yang, H. Transcriptome Profiling Revealed Basis for Growth Heterosis in Hybrid Tilapia (Oreochromis niloticus ♀ × O. aureus ♂). Fishes 2022, 7, 43. [Google Scholar] [CrossRef]
- Roh, H.; Kim, A.; Kim, N.; Lee, Y.; Kim, D.H. Multi-Omics Analysis Provides Novel Insight into Immuno-Physiological Pathways and Development of Thermal Resistance in Rainbow Trout Exposed to Acute Thermal Resistance. Int. J. Mol. Sci. 2020, 21, 9198. [Google Scholar] [CrossRef]
- Chen, Y.J.; Wu, H.; Shen, X.Z. The Ubiquitin–Proteasome System and Its Potential Application in Hepatocellular Carcinoma Therapy. Cancer Lett. 2016, 379, 245–252. [Google Scholar] [CrossRef]
- Peng, J.; Li, W.B.; Wang, B.; Zhang, S.; Xiao, Y.; Han, F.; Wang, Z.Y. UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas plecoglossicida in Large Yellow Croaker (Larimichthys crocea). Int. J. Mol. Sci. 2022, 23, 8298. [Google Scholar] [CrossRef]
- Groettrup, M.; Pelzer, C.; Schmidtke, G.; Hofmann, K. Activating the Ubiquitin Family: UBA6 Challenges the Field. Trends Biochem. Sci. 2008, 33, 230–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.L.; Zhao, L.L.; Wu, H.; Liu, Q.; Liao, L.; Luo, J.; Lian, W.Q.; Cui, C.; Jin, L.; Ma, J.D.; et al. Acute Hypoxia Changes the Mode of Glucose and Lipid Utilization in the Liver of the Largemouth Bass (Micropterus salmoides). Sci. Total Environ. 2020, 713, 135157. [Google Scholar] [CrossRef]
- López Patiño, M.A.; Hernández Pérez, J.; Gesto, M.; Librán Pérez, M.; Míguez, J.M.; Soengas, J.L. Short-Term Time Course of Liver Metabolic Response to Acute Handling Stress in Rainbow Trout, Oncorhynchus mykiss. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 168, 40–49. [Google Scholar] [CrossRef]
- Wang, Y.L.; Meng, R.; Xu, X.R.; Liao, K.; Ran, Z.S.; Xu, J.L.; Cao, J.Y.; Wang, Y.J.; Wang, D.L.; Xu, S.L.; et al. Effects of Nutritional Status and Diet Composition on Fatty Acid Transporters Expression in Zebrafish (Danio rerio). Aquac. Res. 2019, 50, 904–914. [Google Scholar] [CrossRef]
- Bayır, M.; Bayır, A.; Wright, J.M. Divergent Spatial Regulation of Duplicated Fatty Acid-Binding Protein (Fabp) Genes in Rainbow Trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. Part D Genom. Proteom. 2015, 14, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Cheema, S.K.; Agellon, L.B. The Murine and Human Cholesterol 7alpha-Hydroxylase Gene Promoters Are Differentially Responsive to Regulation by Fatty Acids Mediated via Peroxisome Proliferator-Activated Receptor Alpha. J. Biol. Chem. 2000, 275, 12530–12536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ameer, F.; Munir, R.; Zaidi, N. Lipid Metabolism. In Encyclopedia of Cancer, 3rd ed.; Boffetta, P., Hainaut, P., Eds.; Academic Press: Oxford, UK, 2019; pp. 369–373. ISBN 978-0-12-812485-7. [Google Scholar]
- Lu, X.Y.; Shi, X.J.; Hu, A.; Wang, J.Q.; Ding, Y.; Jiang, W.; Sun, M.; Zhao, X.L.; Luo, J.; Qi, W.; et al. Feeding Induces Cholesterol Biosynthesis via the mTORC1–USP20–HMGCR Axis. Nature 2020, 588, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Glencross, B.D.; Santis, C.D.; Bicskei, B.; Taggart, J.B.; Bron, J.E.; Betancor, M.B.; Tocher, D.R. A Comparative Analysis of the Response of the Hepatic Transcriptome to Dietary Docosahexaenoic Acid in Atlantic Salmon (Salmo salar) Post-Smolts. BMC Genom. 2015, 16, 684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.; Stevenson, J.; Kristiana, I.; Brown, A.J. Cholesterol-Dependent Degradation of Squalene Monooxygenase, a Control Point in Cholesterol Synthesis beyond HMG-CoA Reductase. Cell Metab. 2011, 13, 260–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.S.; Li, L.; Li, W.; Yang, F.; Zhang, Z.X.; Liu, Z.Z.; Du, W.J. P53 Transcriptionally Regulates SQLE to Repress Cholesterol Synthesis and Tumor Growth. EMBO Rep. 2021, 22, e52537. [Google Scholar] [CrossRef]
- Gatticchi, L.; de las Heras, J.I.; Sivakumar, A.; Zuleger, N.; Roberti, R.; Schirmer, E.C. Tm7sf2 Disruption Alters Radial Gene Positioning in Mouse Liver Leading to Metabolic Defects and Diabetes Characteristics. Front. Cell Dev. Biol. 2020, 8, 592573. [Google Scholar] [CrossRef]
- Lewinska, M.; Juvan, P.; Perse, M.; Jeruc, J.; Kos, S.; Lorbek, G.; Urlep, Z.; Keber, R.; Horvat, S.; Rozman, D. Hidden Disease Susceptibility and Sexual Dimorphism in the Heterozygous Knockout of Cyp51 from Cholesterol Synthesis. PLoS ONE 2014, 9, e112787. [Google Scholar] [CrossRef] [Green Version]
- Hui, D.Y.; Howles, P.N. Carboxyl Ester Lipase. J. Lipid Res. 2002, 43, 2017–2030. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Liang, Y.S.; Gu, Y.; Yao, F.C.; Liu, Y.F.; Zhang, K.X.; Song, F.B.; Sun, J.L.; Luo, J. Different Reoxygenation Rates Induce Different Metabolic, Apoptotic and Immune Responses in Golden Pompano (Trachinotus blochii) after Hypoxic Stress. Fish Shellfish. Immunol. 2023, 135, 108640. [Google Scholar] [CrossRef]
- Zhao, T.T.; Ma, A.J.; Huang, Z.H.; Liu, Z.F.; Sun, Z.B.; Zhu, C.Y.; Yang, J.K.; Li, Y.D.; Wang, Q.M.; Qiao, X.W.; et al. Transcriptome Analysis Reveals That High Temperatures Alter Modes of Lipid Metabolism in Juvenile Turbot (Scophthalmus maximus) Liver. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100887. [Google Scholar] [CrossRef] [PubMed]
- Gornati, R.; Papis, E.; Rimoldi, S.; Chini, V.; Terova, G.; Prati, M.; Saroglia, M.; Bernardini, G. Molecular Markers for Animal Biotechnology: Sea Bass (Dicentrarchus labrax, L.) HMG-CoA Reductase mRNA. Gene 2005, 344, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Montero, D.; Izquierdo, M.; Tort, L.; Robaina, L.; Vergara, J.M. High Stocking Density Produces Crowding Stress Altering Some Physiological and Biochemical Parameters in Gilthead Seabream, Sparus aurata, Juveniles. Fish Physiol. Biochem. 1999, 20, 53–60. [Google Scholar] [CrossRef]
- Jia, R.; Wang, L.; Hou, Y.R.; Feng, W.R.; Li, B.; Zhu, J. Effects of Stocking Density on the Growth Performance, Physiological Parameters, Redox Status and Lipid Metabolism of Micropterus salmoides in Integrated Rice–Fish Farming Systems. Antioxidants 2022, 11, 1215. [Google Scholar] [CrossRef]
- Raposo de Magalhães, C.; Schrama, D.; Nakharuthai, C.; Boonanuntanasarn, S.; Revets, D.; Planchon, S.; Kuehn, A.; Cerqueira, M.; Carrilho, R.; Farinha, A.P.; et al. Metabolic Plasticity of Gilthead Seabream under Different Stressors: Analysis of the Stress Responsive Hepatic Proteome and Gene Expression. Front. Mar. Sci. 2021, 8, 676189. [Google Scholar] [CrossRef]
- Sivanand, S.; Rhoades, S.; Jiang, Q.Q.; Lee, J.V.; Benci, J.; Zhang, J.W.; Yuan, S.; Viney, I.; Zhao, S.; Carrer, A.; et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol. Cell 2017, 67, 252–265. [Google Scholar] [CrossRef] [Green Version]
- Corominas-Faja, B.; Cuyàs, E.; Gumuzio, J.; Bosch-Barrera, J.; Leis, O.; Martin, Á.G.; Menendez, J.A. Chemical Inhibition of Acetyl-CoA Carboxylase Suppresses Self-Renewal Growth of Cancer Stem Cells. Oncotarget 2014, 5, 8306–8316. [Google Scholar] [CrossRef] [Green Version]
- Cronan, J.E.; Waldrop, G.L. Multi-Subunit Acetyl-CoA Carboxylases. Prog. Lipid Res. 2002, 41, 407–435. [Google Scholar] [CrossRef]
- Bláhová, Z.; Harvey, T.N.; Pšenička, M.; Mráz, J. Assessment of Fatty Acid Desaturase (Fads2) Structure-Function Properties in Fish in the Context of Environmental Adaptations and as a Target for Genetic Engineering. Biomolecules 2020, 10, 206. [Google Scholar] [CrossRef] [Green Version]
- de Lima Luna, A.C.; Forti, F.L. Modulation of SCD1 Activity in Hepatocyte Cell Lines: Evaluation of Genomic Stability and Proliferation. Mol. Cell. Biochem. 2021, 476, 3393–3405. [Google Scholar] [CrossRef]
- Lehner, R.; Quiroga, A.D. Fatty Acid Handling in Mammalian Cells. In Biochemistry of Lipids, Lipoproteins and Membranes, 7th ed.; Ridgway, N.D., McLeod, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 161–200. ISBN 978-0-12-824048-9. [Google Scholar]
- Berthelot, K.; Estevez, Y.; Deffieux, A.; Peruch, F. Isopentenyl Diphosphate Isomerase: A Checkpoint to Isoprenoid Biosynthesis. Biochimie 2012, 94, 1621–1634. [Google Scholar] [CrossRef]
- Neufeld, E.B.; Demosky, S.J.; Stonik, J.A.; Combs, C.; Remaley, A.T.; Duverger, N.; Santamarina-Fojo, S.; Brewer, H.B. The ABCA1 Transporter Functions on the Basolateral Surface of Hepatocytes. Biochem. Biophys. Res. Commun. 2002, 297, 974–979. [Google Scholar] [CrossRef]
- Pickering, A.D.; Pottinger, T.G. Stress Responses and Disease Resistance in Salmonid Fish: Effects of Chronic Elevation of Plasma Cortisol. Fish Physiol. Biochem. 1989, 7, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Hoseini, S.M.; Abtahi, B.; Yousefi, M. Effects of Fasting on Metabolic and Immunological Responses of Common Carp (Cyprinus carpio) to a Further Acute Stress. Aquac. Res. 2019, 50, 1177–1185. [Google Scholar] [CrossRef]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs Are a Unique Set of Fatty Acid Regulated Transcription Factors Controlling Both Lipid Metabolism and Inflammation. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2011, 1812, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Nakajima, T.; Gonzalez, F.J.; Tanaka, N. PPARs as Metabolic Regulators in the Liver: Lessons from Liver-Specific PPAR-Null Mice. Int. J. Mol. Sci. 2020, 21, 2061. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, X.; Liu, Y.; Fang, S.Y.; Wu, Z.J.; Han, C.; Shi, W.Y.; Bao, Y.Z. Effects of Early Florfenicol Exposure on Glutathione Signaling Pathway and PPAR Signaling Pathway in Chick Liver. Ecotoxicol. Environ. Saf. 2022, 237, 113529. [Google Scholar] [CrossRef]
- Xu, H.G.; Zhang, X.; Wei, Y.L.; Sun, B.; Jia, L.L.; Liang, M.Q. Effects of Dietary Phosphorus Level and Stocking Density on Tiger Puffer Takifugu rubripes: Growth Performance, Body Composition, Lipid Metabolism, Deposition of Phosphorus and Calcium, Serum Biochemical Parameters, and Phosphorus Excretion. Aquaculture 2020, 529, 735709. [Google Scholar] [CrossRef]
- Cao, X.F.; Liu, W.B.; Ai, Q.H.; Li, X.S.; Li, J.B.; Fang, W.; Huang, Y.Y.; Wang, C.C.; Jiang, G.Z. High-fat Diet-induced Inflammation Aggravates Hepatic Steatosis of Blunt Snout Bream (Megalobrama amblycephala) through the Transcription Regulation of Fatty Acid Synthesis and Oxidation. Aquacult. Nutr. 2020, 26, 1493–1504. [Google Scholar] [CrossRef]
- Xu, C.; Li, E.; Liu, S.; Huang, Z.P.; Qin, J.G.; Chen, L.Q. Effects of α-Lipoic Acid on Growth Performance, Body Composition, Antioxidant Status and Lipid Catabolism of Juvenile Chinese Mitten Crab Eriocheir sinensis Fed Different Lipid Percentage. Aquaculture 2018, 484, 286–292. [Google Scholar] [CrossRef] [Green Version]
- Anders, N.; Roth, B.; Breen, M. Physiological Response and Survival of Atlantic Mackerel Exposed to Simulated Purse Seine Crowding and Release. Conserv. Physiol. 2021, 9, coab076. [Google Scholar] [CrossRef]
- Dhabhar, F.S. The Power of Positive Stress—A Complementary Commentary. Stress 2019, 22, 526–529. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Dixon, B. Understanding Acute Stress-Mediated Immunity in Teleost Fish. Fish Shellfish. Immunol. Rep. 2021, 2, 100010. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.L.; Sun, J.L.; Liang, J.; Liu, Q.; Luo, J.; Li, Z.Q.; Yan, T.M.; Zhou, J.; Yang, S. Enhancing Lipid Metabolism and Inducing Antioxidant and Immune Responses to Adapt to Acute Hypoxic Stress in Schizothorax prenanti. Aquaculture 2020, 519, 734933. [Google Scholar] [CrossRef]
- Okoye, C.N.; Chinnappareddy, N.; Stevens, D.; Kamunde, C. Factors Affecting Liver Mitochondrial Hydrogen Peroxide Emission. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2022, 259, 110713. [Google Scholar] [CrossRef] [PubMed]
- Cannito, S.; Novo, E.; Parola, M. Therapeutic Pro-Fibrogenic Signaling Pathways in Fibroblasts. Adv. Drug Deliv. Rev. 2017, 121, 57–84. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, K.C.; Pan, J.M.; Guo, H.Y.; Liu, B.S.; Zhang, N.; Yang, J.W.; Zhang, D.C. Characterization of the MMP9 Gene and Its Association with Cryptocaryon irritans Resistance Traits in Trachinotus ovatus (Linnaeus, 1758). Genes 2023, 14, 475. [Google Scholar] [CrossRef]
- Kluger, M.A.; Zahner, G.; Paust, H.-J.; Schaper, M.; Magnus, T.; Panzer, U.; Stahl, R.A.K. Leukocyte-Derived MMP9 Is Crucial for the Recruitment of Proinflammatory Macrophages in Experimental Glomerulonephritis. Kidney Int. 2013, 83, 865–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.P.; Deng, F.; Sharma, I.; Kanwar, Y.S. Myo-Inositol Oxygenase Overexpression Exacerbates Cadmium-Induced Kidney Injury via Oxidant Stress and Necroptosis. Am. J. Physiol. Ren. Physiol. 2022, 322, F344–F359. [Google Scholar] [CrossRef]
- Yoshihara, D.; Fujiwara, N.; Ookawara, T.; Kato, S.; Sakiyama, H.; Yokoe, S.; Eguchi, H.; Suzuki, K. Protective Role of Glutathione S-Transferase A4 Induced in Copper/Zinc-Superoxide Dismutase Knockout Mice. Free. Radic. Biol. Med. 2009, 47, 559–567. [Google Scholar] [CrossRef]
- Ramming, T.; Hansen, H.G.; Nagata, K.; Ellgaard, L.; Appenzeller-Herzog, C. GPx8 Peroxidase Prevents Leakage of H2O2 from the Endoplasmic Reticulum. Free. Radic. Biol. Med. 2014, 70, 106–116. [Google Scholar] [CrossRef]
- Berry, F.B.; Skarie, J.M.; Mirzayans, F.; Fortin, Y.; Hudson, T.J.; Raymond, V.; Link, B.A.; Walter, M.A. FOXC1 Is Required for Cell Viability and Resistance to Oxidative Stress in the Eye through the Transcriptional Regulation of FOXO1A. Hum. Mol. Genet. 2008, 17, 490–505. [Google Scholar] [CrossRef] [Green Version]
- Ortuño, J.; Esteban, M.A.; Meseguer, J. Effects of Short-Term Crowding Stress on the Gilthead Seabream (Sparus aurata L.) Innate Immune Response. Fish Shellfish. Immunol. 2001, 11, 187–197. [Google Scholar] [CrossRef]
- Huang, X.J.; Xu, Y.L.; Yang, Q.Y.; Chen, J.Q.; Zhang, T.; Li, Z.C.; Guo, C.P.; Chen, H.; Wu, H.; Li, N. Efficacy and Biological Safety of Lopinavir/Ritonavir Based Anti-Retroviral Therapy in HIV-1-Infected Patients: A Meta-Analysis of Randomized Controlled Trials. Sci. Rep. 2015, 5, 8528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, M.; Kommaly, O.; Li, D.P.; Wang, H.S.; Liang, X.; Tang, R.; Li, L.; Zhang, X.; Chi, W. Effect of acute crowding stress on the ubiquitin-proteasome system in the muscle of grass carp (Ctenopharyngodon idellus). J. Fish. Sci. China 2019, 26, 427. [Google Scholar] [CrossRef]
- Yada, T.; Tort, L. Stress and Disease Resistance: Immune System and Immunoendocrine Interactions. In Fish Physiology; Schreck, C.B., Tort, L., Farrell, A.P., Brauner, C.J., Eds.; Biology of Stress in Fish; Academic Press: Cambridge, MA, USA, 2016; Volume 35, pp. 365–403. [Google Scholar]
- Huo, H.H.; Gao, X.Q.; Fei, F.; Qin, F.; Huang, B.; Liu, B.L. Transcriptomic Profiling of the Immune Response to Crowding Stress in Juvenile Turbot (Scophthalmus maximus). J. Ocean. Univ. China 2020, 19, 911–922. [Google Scholar] [CrossRef]
- Jia, R.; Liu, B.L.; Feng, W.R.; Han, C.; Huang, B.; Lei, J.L. Stress and Immune Responses in Skin of Turbot (Scophthalmus maximus) under Different Stocking Densities. Fish Shellfish. Immunol. 2016, 55, 131–139. [Google Scholar] [CrossRef]
- Ellison, A.R.; Uren Webster, T.M.; Rodriguez-Barreto, D.; de Leaniz, C.G.; Consuegra, S.; Orozco ter Wengel, P.; Cable, J. Comparative Transcriptomics Reveal Conserved Impacts of Rearing Density on Immune Response of Two Important Aquaculture Species. Fish Shellfish. Immunol. 2020, 104, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Yarahmadi, P.; Miandare, H.K.; Fayaz, S.; Caipang, C.M.A. Increased Stocking Density Causes Changes in Expression of Selected Stress- and Immune-Related Genes, Humoral Innate Immune Parameters and Stress Responses of Rainbow Trout (Oncorhynchus mykiss). Fish Shellfish. Immunol. 2016, 48, 43–53. [Google Scholar] [CrossRef]
- Haase, D.; Rieger, J.K.; Witten, A.; Stoll, M.; Bornberg-Bauer, E.; Kalbe, M.; Schmidt-Drewello, A.; Scharsack, J.P.; Reusch, T.B.H. Comparative Transcriptomics of Stickleback Immune Gene Responses upon Infection by Two Helminth Parasites, Diplostomum pseudospathaceum and Schistocephalus solidus. Zoology 2016, 119, 307–313. [Google Scholar] [CrossRef]
- Wang, T.; Liang, J.; Xiang, X.; Yuan, J.; Chen, X.; Xiang, X.; Yang, J. Functional Identification and Expressional Responses of Large Yellow Croaker (Larimichthys crocea) Interleukin-8 and Its Receptor. Fish Shellfish. Immunol. 2019, 87, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Shariat, M.; Heydrzadeh, M.; Abolhassani, H.; Bemanian, M.; Yazdani, R. Complement Deficiencies. In Inborn Errors of Immunity; Academic Press: Cambridge, MA, USA, 2021; pp. 291–315. ISBN 978-0-12-821028-4. [Google Scholar]
- Sarma, J.V.; Ward, P.A. The Complement System. Cell Tissue Res. 2011, 343, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Arlaud, G.J.; Gaboriaud, C.; Thielens, N.M.; Budayova-Spano, M.; Rossi, V.; Fontecilla Camps, J.C. Structural Biology of the C1 Complex of Complement Unveils the Mechanisms of Its Activation and Proteolytic Activity. Mol. Immunol. 2002, 39, 383–394. [Google Scholar] [CrossRef]
- Lin, W.; Li, L.; Chen, J.; Li, D.P.; Hou, J.; Guo, H.H.; Shen, J.Z. Long-Term Crowding Stress Causes Compromised Nonspecific Immunity and Increases Apoptosis of Spleen in Grass Carp (Ctenopharyngodon idella). Fish Shellfish. Immunol. 2018, 80, 540–545. [Google Scholar] [CrossRef] [PubMed]








| Gene Name | Primer Sequence (5′→3′) | Annealing Temperature/°C |
|---|---|---|
| thbs1 | F: ATCCTGGCATTGCTGTTGGTTAC | 56 |
| R: CGTCTCTGTCCGTGTTGATGAAG | ||
| mr1 | F: GAACGAGGACATCATCACCAGAC | 56 |
| R: ATCCAGCAGCACCGATAAGAATAAG | ||
| cela2a | F: TGTTACACTGATGGAGCCTGGAG | 56 |
| R: GGTGAAGACGGTGGGTTTGTTC | ||
| fabp1 | F: CCTGGAGACGATCACTGGAGAG | 56 |
| R: GCCACCGACAGTCATAGTATTGC | ||
| fads2 | F: GCCACCTGTCTGTCTTCAAGAAG | 55 |
| R: AATGCCGATGATTCCACCAGTTG | ||
| acaca | F: TCAACAAGAAGTCAGAGCGAGAAG | 54 |
| R: CACGGATGCCACTGCGATAC | ||
| fps | F: TGGAGGCAAGCGGAACAGAG | 58 |
| R: ACCAGCCGACCAACAGAGC | ||
| cyp51 | F: TCGGGAGCACATTCACCTACC | 58 |
| R: CTGGAGTGGTTAGCCTGGAGTAG | ||
| acly | F: GTGATGGGAGAAGTTGGCAAGAC | 59 |
| R: AGGAGGAAGTTGGCAGTGTGAG | ||
| β-actin | F: CGACGGTCAGGTCATCAC | 56 |
| R: GCCAGCAGACTCCATTCC |
| Sample | Raw_Data (bp) | Clean_Data (bp) | Q20 | Q30 | GC |
|---|---|---|---|---|---|
| AL-1 | 6,265,646,400 | 6,151,585,554 | 98.42% | 95.21% | 51.46% |
| AL-2 | 7,541,598,300 | 7,388,812,971 | 98.07% | 93.91% | 51.33% |
| AL-3 | 7,264,445,700 | 7,157,111,386 | 98.00% | 93.92% | 51.16% |
| AH-1 | 8,141,713,200 | 7,991,147,520 | 97.65% | 92.71% | 50.46% |
| AH-2 | 6,561,749,100 | 6,474,655,672 | 98.48% | 95.39% | 50.47% |
| AH-3 | 8,086,893,900 | 7,960,620,927 | 97.57% | 92.75% | 50.88% |
| CL-1 | 7,398,075,900 | 7,255,941,269 | 97.94% | 93.53% | 50.69% |
| CL-2 | 7,779,589,800 | 7,611,675,168 | 98.15% | 94.12% | 50.30% |
| CL-3 | 6,654,809,100 | 6,544,452,925 | 98.39% | 95.06% | 50.17% |
| CH-1 | 6,395,992,200 | 6,273,751,676 | 98.39% | 95.13% | 50.75% |
| CH-2 | 6,636,990,600 | 6,532,623,483 | 98.52% | 95.46% | 51.12% |
| CH-3 | 7,088,955,300 | 6,964,895,805 | 98.37% | 95.11% | 50.27% |
| Sample | Clean_Data | Unique_Mapped | Total_Mapped | Exon |
|---|---|---|---|---|
| AL-1 | 41,495,774 | 77.65% | 86.30% | 70.87% |
| AL-2 | 49,892,258 | 76.97% | 85.85% | 70.61% |
| AL-3 | 48,103,404 | 76.92% | 85.82% | 70.39% |
| AH-1 | 53,924,360 | 75.97% | 84.13% | 69.83% |
| AH-2 | 43,494,574 | 76.77% | 85.07% | 69.89% |
| AH-3 | 53,560,720 | 76.43% | 84.78% | 70.76% |
| CL-1 | 48,972,544 | 76.51% | 85.67% | 66.96% |
| CL-2 | 51,557,256 | 77.00% | 85.71% | 65.92% |
| CL-3 | 44,080,604 | 77.69% | 86.16% | 65.35% |
| CH-1 | 42,312,210 | 75.96% | 85.20% | 66.26% |
| CH-2 | 43,936,880 | 76.07% | 85.79% | 67.07% |
| CH-3 | 46,941,622 | 76.11% | 85.20% | 65.05% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Jia, C.; Meng, Q.; Xu, D.; Zhang, Z.; Zhu, F.; Zhao, Y.; Sun, R.; Yang, Y.; Chen, S. Transcriptome-Based Analysis of the Liver Response Mechanism of Black Porgy (Acanthopagrus schlegelii) to Stocking Density. Fishes 2023, 8, 356. https://doi.org/10.3390/fishes8070356
Zhou T, Jia C, Meng Q, Xu D, Zhang Z, Zhu F, Zhao Y, Sun R, Yang Y, Chen S. Transcriptome-Based Analysis of the Liver Response Mechanism of Black Porgy (Acanthopagrus schlegelii) to Stocking Density. Fishes. 2023; 8(7):356. https://doi.org/10.3390/fishes8070356
Chicago/Turabian StyleZhou, Tangjian, Chaofeng Jia, Qian Meng, Dafeng Xu, Zhiwei Zhang, Fei Zhu, Yonglei Zhao, Ruijian Sun, Yunxia Yang, and Shuyin Chen. 2023. "Transcriptome-Based Analysis of the Liver Response Mechanism of Black Porgy (Acanthopagrus schlegelii) to Stocking Density" Fishes 8, no. 7: 356. https://doi.org/10.3390/fishes8070356
APA StyleZhou, T., Jia, C., Meng, Q., Xu, D., Zhang, Z., Zhu, F., Zhao, Y., Sun, R., Yang, Y., & Chen, S. (2023). Transcriptome-Based Analysis of the Liver Response Mechanism of Black Porgy (Acanthopagrus schlegelii) to Stocking Density. Fishes, 8(7), 356. https://doi.org/10.3390/fishes8070356