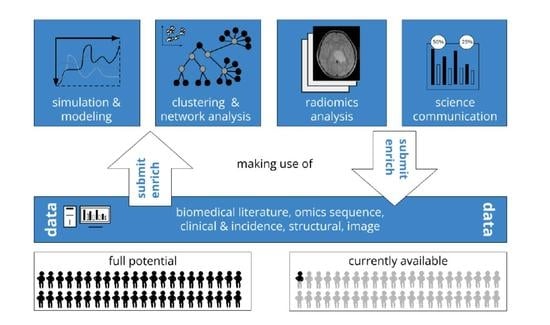
Open Data to Support CANCER Science—A Bioinformatics Perspective on Glioma Research
Abstract
:Simple Summary
Abstract
1. Introduction
2. Open Data for Cancer Research
- Why open data research?—from open to FAIR (findable, accessible, interoperable, and reusable).
- General biomedical data providers.
- Cancer specific data initiatives and resources.
- Metadata for AI in cancer research.
- Explainability and causability.
- Fostering exchange and use cases for glioma research.
2.1. Ad 1. Why Open Data Research?
2.2. General Biomedical Data Providers
2.3. Cancer Specific Data Initiatives and Resources
2.4. Metadata for AI in Cancer Research
2.5. Explainability and Causability
2.6. Fostering Exchange and Use Cases for Glioma Research
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CCDS | Consensus Coding Sequence Database |
| CCMI | Cancer Cell Map Initiative |
| CGGA | Chinese Glioma Genome Atlas |
| COSMIC | Catalogue of Somatic Mutations in Cancer |
| DKTK | German Cancer Consortium: Deutsches Konsortium für Translationale Krebsforschung |
| EMBL–EBI | European Molecular Biology Laboratory - European Bioinformatics Institute |
| FAIR | Findable, Accessible, Interoperable, and Reusable |
| GA4GH | Global Alliance for Genomics and Health |
| GDC | Genomic Data Commons |
| IARC | International Agency for Research on Cancer |
| ICGC | International Cancer Genome Consortium |
| ICSU | International Council of Scientific Unioins |
| ISC | International Science Counsil |
| ISSC | International Social Science Counsil |
| NASA | National Aeronautics and Space Administration |
| NCBI | National Center for Biotechnology Information |
| NCI | National Cancer Institute |
| PPI | Protein Protein Interaction |
| PCAWG | Pancancer Analysis of Whole Genomes |
| PCGP | Pediatric Cancer Genome Project |
| PDB(e) | Protein Data Bank (in Europe) |
| PMC | PubMed Central |
| PPI | protein-protein interaction |
| SEER | Surveillance, Epidemiology, and End Results |
| TARGET | Therapeutically Applicable Research to Generate Effective Treatments |
| TCGA | The Cancer Genome Project |
| TCIA | The Cancer Imaging Archive |
| UCSC | University of California Santa Cruz |
| WHO | World Health Organization |
| WSI | Wellcome Sanger Institute |
| xAI | Explainable Artificial Intelligence |
References
- Jean-Quartier, C.; Jeanquartier, F.; Jurisica, I.; Holzinger, A. In silico cancer research towards 3R. BMC Cancer 2018, 18, 408. [Google Scholar] [CrossRef]
- Zuiderwijk, A.; Shinde, R.; Jeng, W. What drives and inhibits researchers to share and use open research data? A systematic literature review to analyze factors influencing open research data adoption. PLoS ONE 2020, 15, e0239283. [Google Scholar] [CrossRef]
- Vamathevan, J.; Apweiler, R.; Birney, E. Biomolecular data resources: Bioinformatics infrastructure for biomedical data science. Annu. Rev. Biomed. Data Sci. 2019, 2, 199–222. [Google Scholar] [CrossRef]
- Aronova, E.; Baker, K.S.; Oreskes, N. Big science and big data in biology: From the international geophysical year through the international biological program to the long term ecological research (LTER) Network, 1957—-Present. Hist. Stud. Nat. Sci. 2010, 40, 183–224. [Google Scholar] [CrossRef]
- Esteban, M.J.; Puppo, G. The New International Science Council–A Global Voice for Science. EMS Newsl. 2018, 109, 49. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, B.; Kemmerer, S.; Parks, C. A Brief History of Early Product Data Exchange Standards; NIST Interagency/Internal Report (NISTIR); National Institute of Standards and Technology: Gaithersburg, MD, USA, 1998. [Google Scholar]
- Nicol, A.; Caruso, J.; Archambault, É. Open data access policies and strategies in the European research area and beyond. Info@ Sci. 2013, 1, 495–6505. [Google Scholar]
- National Research Council. On the Full and Open Exchange of Scientific Data; The National Academies: Washington, DC, USA, 1995. [Google Scholar] [CrossRef]
- Hinkson, I.V.; Davidsen, T.M.; Klemm, J.D.; Chandramouliswaran, I.; Kerlavage, A.R.; Kibbe, W.A. A comprehensive infrastructure for big data in cancer research: Accelerating cancer research and precision medicine. Front. Cell Dev. Biol. 2017, 5, 83. [Google Scholar] [CrossRef]
- Milius, D.; Dove, E.S.; Chalmers, D.; Dyke, S.O.; Kato, K.; Nicolas, P.; Ouellette, B.F.; Ozenberger, B.; Rodriguez, L.L.; Zeps, N.; et al. The International Cancer Genome Consortium’s evolving data-protection policies. Nat. Biotechnol. 2014, 32, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Joos, S.; Nettelbeck, D.M.; Reil-Held, A.; Engelmann, K.; Moosmann, A.; Eggert, A.; Hiddemann, W.; Krause, M.; Peters, C.; Schuler, M.; et al. German Cancer Consortium (DKTK)–A national consortium for translational cancer research. Mol. Oncol. 2019, 13, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Lawler, M.; Siu, L.L.; Rehm, H.L.; Chanock, S.J.; Alterovitz, G.; Burn, J.; Calvo, F.; Lacombe, D.; Teh, B.T.; North, K.N.; et al. All the world’s a stage: Facilitating discovery science and improved cancer care through the global alliance for genomics and health. Cancer Discov. 2015, 5, 1133–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- OECD. Making Open Science a Reality; OECD Science, Technology and Industry Policy Papers; OECD: Paris, France, 2015; Volume 25. [Google Scholar]
- Besançon, L.; Peiffer-Smadja, N.; Segalas, C.; Jiang, H.; Masuzzo, P.; Smout, C.; Billy, E.; Deforet, M.; Leyrat, C. Open science saves lives: Lessons from the COVID-19 pandemic. BMC Med. Res. Methodol. 2021, 21, 117. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansone, S.A.; McQuilton, P.; Rocca-Serra, P.; Gonzalez-Beltran, A.; Izzo, M.; Lister, A.L.; Thurston, M. FAIRsharing as a community approach to standards, repositories and policies. Nat. Biotechnol. 2019, 37, 358–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centre, D.C. Disciplinary Metadata. Available online: https://www.dcc.ac.uk/guidance/standards/metadata (accessed on 12 December 2021).
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2019, 47, D23. [Google Scholar] [CrossRef] [Green Version]
- Sarkans, U.; Füllgrabe, A.; Ali, A.; Athar, A.; Behrangi, E.; Diaz, N.; Fexova, S.; George, N.; Iqbal, H.; Kurri, S.; et al. From ArrayExpress to BioStudies. Nucleic Acids Res. 2021, 49, D1502–D1506. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The single global macromolecular structure archive. In Protein Crystallography; Humana Press: New York, NY, USA, 2017; pp. 627–641. [Google Scholar]
- Pujar, S.; O’Leary, N.A.; Farrell, C.M.; Loveland, J.E.; Mudge, J.M.; Wallin, C.; Girón, C.G.; Diekhans, M.; Barnes, I.; Bennett, R.; et al. Consensus coding sequence (CCDS) database: A standardized set of human and mouse protein-coding regions supported by expert curation. Nucleic Acids Res. 2018, 46, D221–D228. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A comprehensive resource with functional genomic data from Chinese gliomas. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Sarkans, U.; Chiu, W.; Collinson, L.; Darrow, M.C.; Ellenberg, J.; Grunwald, D.; Hériché, J.K.; Iudin, A.; Martins, G.G.; Meehan, T.; et al. REMBI: Recommended Metadata for Biological Images—enabling reuse of microscopy data in biology. Nat. Methods 2021, 18, 1418–1422. [Google Scholar] [CrossRef]
- Touré, V.; Flobak, Å.; Niarakis, A.; Vercruysse, S.; Kuiper, M. The status of causality in biological databases: Data resources and data retrieval possibilities to support logical modeling. Briefings Bioinform. 2021, 22, bbaa390. [Google Scholar] [CrossRef]
- Kingsley, J.L.; Costello, J.R.; Raghunand, N.; Rejniak, K.A. Bridging cell-scale simulations and radiologic images to explain short-time intratumoral oxygen fluctuations. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Hormuth, D.A.; Al Feghali, K.A.; Elliott, A.M.; Yankeelov, T.E.; Chung, C. Image-based personalization of computational models for predicting response of high-grade glioma to chemoradiation. Sci. Rep. 2021, 11, 8520. [Google Scholar] [CrossRef]
- Jeanquartier, F.; Jean-Quartier, C.; Cemernek, D.; Holzinger, A. In silico modeling for tumor growth visualization. BMC Syst. Biol. 2016, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, H.; Schirner, M.; Dhollander, T.; Jeurissen, B.; Achten, E.; Van Roost, D.; Ritter, P.; Marinazzo, D. Modeling brain dynamics after tumor resection using The Virtual Brain. Neuroimage 2020, 213, 116738. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, N.; Delbridge, C.; Gempt, J.; Feuchtinger, A.; Walch, A.; Schirmer, L.; Bunk, W.; Aschenbrenner, T.; Liesche-Starnecker, F.; Schlegel, J. The intratumoral heterogeneity reflects the intertumoral subtypes of glioblastoma multiforme: A regional immunohistochemistry analysis. Front. Oncol. 2020, 10, 494. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, D.; Liu, L.; Li, Z.; Zuo, L.; Zhou, L.; Du, Q.; Jing, Z.; Zhang, X.; Sun, Z. Integrative Analysis of Metabolomic and Transcriptomic Data Reveals Metabolic Alterations in Glioma Patients. J. Proteome Res. 2021, 20, 2206–2215. [Google Scholar] [CrossRef]
- Yang, Y.; Sui, Y.; Xie, B.; Qu, H.; Fang, X. GliomaDB: A web server for integrating glioma omics data and interactive analysis. Genom. Proteom. Bioinform. 2019, 17, 465–471. [Google Scholar] [CrossRef]
- Jean-Quartier, C.; Jeanquartier, F.; Holzinger, A. Open data for differential network analysis in glioma. Int. J. Mol. Sci. 2020, 21, 547. [Google Scholar] [CrossRef] [Green Version]
- Jean-Quartier, C.; Jeanquartier, F.; Ridvan, A.; Kargl, M.; Mirza, T.; Stangl, T.; Markaĉ, R.; Jurada, M.; Holzinger, A. Mutation-based clustering and classification analysis reveals distinctive age groups and age-related biomarkers for glioma. BMC Med. Inform. Decis. Mak. 2021, 21, 77. [Google Scholar] [CrossRef]
- Jeanquartier, F.; Jean-Quartier, C.; Holzinger, A. Use case driven evaluation of open databases for pediatric cancer research. BioData Min. 2019, 12, 2. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef] [Green Version]
- Krasnov, G.S.; Kudryavtseva, A.V.; Snezhkina, A.V.; Lakunina, V.A.; Beniaminov, A.D.; Melnikova, N.V.; Dmitriev, A.A. Pan-cancer analysis of TCGA data revealed promising reference genes for qPCR normalization. Front. Genet. 2019, 10, 97. [Google Scholar] [CrossRef] [Green Version]
- Ortmayr, K.; Dubuis, S.; Zampieri, M. Metabolic profiling of cancer cells reveals genome-wide crosstalk between transcriptional regulators and metabolism. Nat. Commun. 2019, 10, 1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.; Li, G.; Zhai, Y.; Huang, L.; Feng, Y.; Wang, D.; Zhang, W.; Hu, H. Redox regulator GLRX is associated with tumor immunity in glioma. Front. Immunol. 2020, 11, 3028. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Tustison, N.J.; Patel, S.H.; Meyer, C.H. Brain tumor segmentation using an ensemble of 3d u-nets and overall survival prediction using radiomic features. Front. Comput. Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Kofler, F.; Berger, C.; Waldmannstetter, D.; Lipkova, J.; Ezhov, I.; Tetteh, G.; Kirschke, J.; Zimmer, C.; Wiestler, B.; Menze, B.H. BraTS Toolkit: Translating BraTS brain tumor segmentation algorithms into clinical and scientific practice. Front. Neurosci. 2020, 14, 125. [Google Scholar] [CrossRef]
- Banerjee, S.; Mitra, S.; Masulli, F.; Rovetta, S. Glioma classification using deep radiomics. SN Comput. Sci. 2020, 1, 209. [Google Scholar] [CrossRef]
- Lu, V.M.; Power, E.A.; Kerezoudis, P.; Daniels, D.J. The 100 most-cited articles about diffuse intrinsic pontine glioma: A bibliometric analysis. Child’s Nerv. Syst. 2019, 35, 2339–2346. [Google Scholar] [CrossRef] [PubMed]
- Akmal, M.; Hasnain, N.; Rehan, A.; Iqbal, U.; Hashmi, S.; Fatima, K.; Farooq, M.Z.; Khosa, F.; Siddiqi, J.; Khan, M.K. Glioblastome multiforme: A bibliometric analysis. World Neurosurg. 2020, 136, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Celiku, O.; Johnson, S.; Zhao, S.; Camphausen, K.; Shankavaram, U. Visualizing molecular profiles of glioblastoma with GBM-BioDP. PLoS ONE 2014, 9, e101239. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Yang, T.H.O.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The immune landscape of cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [Green Version]
- Prior, F.W.; Clark, K.; Commean, P.; Freymann, J.; Jaffe, C.; Kirby, J.; Moore, S.; Smith, K.; Tarbox, L.; Vendt, B.; et al. TCIA: An information resource to enable open science. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 1282–1285. [Google Scholar]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Mazor, T.; De Bruijn, I.; Abeshouse, A.; Baiceanu, D.; Erkoc, Z.; Gross, B.; Higgins, D.; Jagannathan, P.K.; Kalletla, K.; et al. The cBioPortal for Cancer Genomics. Cancer Res. 2021, 81, 207. [Google Scholar]
- Pratt, D.; Chen, J.; Pillich, R.; Rynkov, V.; Gary, A.; Demchak, B.; Ideker, T. NDEx 2.0: A clearinghouse for research on cancer pathways. Cancer Res. 2017, 77, e58–e61. [Google Scholar] [CrossRef] [Green Version]
- Pavlopoulou, A.; Spandidos, D.A.; Michalopoulos, I. Human cancer databases. Oncol. Rep. 2015, 33, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Avsec, Ž.; Kreuzhuber, R.; Israeli, J.; Xu, N.; Cheng, J.; Shrikumar, A.; Banerjee, A.; Kim, D.S.; Beier, T.; Urban, L.; et al. The Kipoi repository accelerates community exchange and reuse of predictive models for genomics. Nat. Biotechnol. 2019, 37, 592–600. [Google Scholar] [CrossRef]
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Matschinske, J.; Alcaraz, N.; Benis, A.; Golebiewski, M.; Grimm, D.G.; Heumos, L.; Kacprowski, T.; Lazareva, O.; List, M.; Louadi, Z.; et al. The AIMe registry for artificial intelligence in biomedical research. Nat. Methods 2021, 18, 1128–1131. [Google Scholar] [CrossRef] [PubMed]
- Kleppe, A.; Skrede, O.J.; De Raedt, S.; Liestøl, K.; Kerr, D.J.; Danielsen, H.E. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 2021, 21, 199–211. [Google Scholar] [CrossRef]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 2020, 28, 104115. [Google Scholar] [CrossRef]
- Commission, E. Proposal for a Regulation of the European Parliament and of the Council Laying down Harmonised Rules on Artificial Intelligence (Artificial Intelligence Act) and Amending Certain Union Legislative Acts. 2021. Available online: https://digital-strategy.ec.europa.eu/en/policies/regulatory-framework-ai (accessed on 12 December 2021).
- Eberle, R.; Kaplan, D.; Montague, R. Hempel and Oppenheim on explanation. Philos. Sci. 1961, 28, 418–428. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef] [Green Version]
- Samek, W.; Montavon, G.; Vedaldi, A.; Hansen, L.K.; Müller, K.R. Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer Nature: Cham, Switzerland, 2019; Volume 11700. [Google Scholar] [CrossRef]
- Holzinger, A.; Malle, B.; Saranti, A.; Pfeifer, B. Towards multi-modal causability with Graph Neural Networks enabling information fusion for explainable AI. Inf. Fusion 2021, 71, 28–37. [Google Scholar] [CrossRef]
- Holzinger, A.; Mueller, H. Toward Human-AI Interfaces to Support Explainability and Causability in Medical AI. IEEE Comput. 2021, 54, 78–86. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Mueller, H. Causability and Explainability of Artificial Intelligence in Medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, A.; Carrington, A.; Mueller, H. Measuring the Quality of Explanations: The System Causability Scale (SCS). Comparing Human and Machine Explanations. KI-Kuenstliche Intell. 2020, 34, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearl, J. Causality: Models, Reasoning, and Inference, 2nd ed.; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Holzinger, A. Explainable ai and multi-modal causability in medicine. i-com 2020, 19, 171–179. [Google Scholar] [CrossRef]
- Wulczyn, E.; Nagpal, K.; Symonds, M.; Moran, M.; Plass, M.; Reihs, R.; Nader, F.; Tan, F.; Cai, Y.; Brown, T.; et al. Predicting Prostate Cancer-Specific Mortality with AI-based Gleason Grading. arXiv 2020, arXiv:2012.05197. [Google Scholar]
- Das, T.; Andrieux, G.; Ahmed, M.; Chakraborty, S. Integration of online omics-data resources for cancer research. Front. Genet. 2020, 11, 578345. [Google Scholar] [CrossRef]
- Wishart, D.S.; Mandal, R.; Stanislaus, A.; Ramirez-Gaona, M. Cancer metabolomics and the human metabolome database. Metabolites 2016, 6, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Heimberger, A.B.; Lu, Z.; Wu, X.; Hodges, T.R.; Song, R.; Shen, J. Metabolomics profiling in plasma samples from glioma patients correlates with tumor phenotypes. Oncotarget 2016, 7, 20486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Diaz, O.; Kushibar, K.; Osuala, R.; Linardos, A.; Garrucho, L.; Igual, L.; Radeva, P.; Prior, F.; Gkontra, P.; Lekadir, K. Data preparation for artificial intelligence in medical imaging: A comprehensive guide to open-access platforms and tools. Phys. Medica 2021, 83, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Shui, L.; Ren, H.; Yang, X.; Li, J.; Chen, Z.; Yi, C.; Zhu, H.; Shui, P. Era of radiogenomics in precision medicine: An emerging approach for prediction of the diagnosis, treatment and prognosis of tumors. Front. Oncol. 2020, 10, 3195. [Google Scholar]
- National Academies of Sciences; Engineering, and Medicine. Communicating Science Effectively: A Research Agenda; National Academies Press: Washington, DC, USA, 2017. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 590–597. [Google Scholar]
- Krogan, N.J.; Lippman, S.; Agard, D.A.; Ashworth, A.; Ideker, T. The cancer cell map initiative: Defining the hallmark networks of cancer. Mol. Cell 2015, 58, 690–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, A.J.; Jin, M.C.; Wu, A.; Bhambhvani, H.P.; Li, G.; Grant, G.A. Risk of secondary neoplasms after external-beam radiation therapy treatment of pediatric low-grade gliomas: A SEER analysis, 1973–2015. J. Neurosurgery Pediatr. 2021, 1, 1–9. [Google Scholar] [CrossRef]
- Kearney, A.; IQVIA. Oncology Data Landscape in Europe Data Sources & Initiatives; Technical Report; efpia: Brussels, Belgium, 2018. [Google Scholar]
- Obermeyer, Z.; Emanuel, E.J. Predicting the future—big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016, 375, 1216. [Google Scholar] [CrossRef] [Green Version]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing medical imaging data for machine learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Marble, H.D.; Huang, R.; Dudgeon, S.N.; Lowe, A.; Herrmann, M.D.; Blakely, S.; Leavitt, M.O.; Isaacs, M.; Hanna, M.G.; Sharma, A.; et al. A regulatory science initiative to harmonize and standardize digital pathology and machine learning processes to speed up clinical innovation to patients. J. Pathol. Inform. 2020, 11, 22. [Google Scholar] [PubMed]
- Cabitza, F.; Zeitoun, J.D. The proof of the pudding: In praise of a culture of real-world validation for medical artificial intelligence. Ann. Transl. Med. 2019, 7, 161. [Google Scholar] [CrossRef] [PubMed]
- Zuiderwijk, A.; de Reuver, M. Why open government data initiatives fail to achieve their objectives: Categorizing and prioritizing barriers through a global survey. Transform. Gov. People Process. Policy 2021, 15, 377–395. [Google Scholar] [CrossRef]

| Name | Type of Data | Provider |
|---|---|---|
| general biomedical data resources | ||
| Pubmed Central (PMC) | publications, references | NCBI (NIH) |
| PubChem | molecule information | NCBI (NIH) |
| Gene Expression Omnibus (GEO) | gene expression | NCBI (NIH) |
| Europe PMC | publications, references | EMBL–EBI |
| Ensembl | genomic information | EMBL–EBI |
| UniProt | protein information | EMBL–EBI |
| Protein Data Bank in Europe (PDBe) | protein structures | EMBL–EBI |
| ChEMBL | molecule information | EMBL–EBI |
| ArrayExpress/Biostudies | from functional genomics to a variety of study data | EMBL–EBI |
| Expression Atlas | gene expression | EMBL–EBI |
| BioImage Archive (BIA) | images of all scales | EMBL–EBI |
| Protein Data Bank (PDB) | protein structures | joint, worldwide |
| Consensus Coding Sequence Database (CCDS) | genome sequences | joint, worldwide |
| cancer-specific resources | ||
| GLOBOCAN | cancer statistics | IARC (WHO) |
| The Cancer Genome Atlas (TCGA) | cancer genomics | NCI (NIH) |
| The Cancer Imaging Archive (TCIA) | cancer images | NCI (NIH) |
| Surveillance, Epidemiology, and End Results (SEER) | cancer incidences | NCI (NIH) |
| International Cancer Genome Consortium (ICGC) | cancer genomics | joint, worldwide |
| Catalogue Of Somatic Mutations In Cancer (COSMIC) | cancer somatic mutations | WSI (England) |
| cBio Cancer Genomics Portal (cBioPortal) | cancer genomics | joint, worldwide |
| Chinese Glioma Genome Atlas (CGGA) | glioma genomics | Beijing Neuro-surgical Institute |
| Pediatric Cancer Genome Project (PCGP) | cancer genomics | joint, worldwide |
| Cancer Cell Map Initiative (CCMI) | cancer cell maps | joint (UCSC a.o.) |
| kipoi | cancer genomic models | joint, worldwide |
| Example USE Cases | Mixed | Open | |
|---|---|---|---|
| modeling and simulation | tumor growth, migration, angiogenesis | [27,28] | [29,30] |
| clustering and network analysis | biomarker discovery, grading, subtype, drug and pathway analysis | [31,32,33] | [34,35,36,37,38,39,40] |
| radiomic analysis | diagnosis and survival | [28,41] | [42,43,44] |
| information retrieval and science communication | epidemiology, education, investigation, bibliometrics | [45,46,47] | [23,48,49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeanquartier, F.; Jean-Quartier, C.; Stryeck, S.; Holzinger, A. Open Data to Support CANCER Science—A Bioinformatics Perspective on Glioma Research. Onco 2021, 1, 219-229. https://doi.org/10.3390/onco1020016
Jeanquartier F, Jean-Quartier C, Stryeck S, Holzinger A. Open Data to Support CANCER Science—A Bioinformatics Perspective on Glioma Research. Onco. 2021; 1(2):219-229. https://doi.org/10.3390/onco1020016
Chicago/Turabian StyleJeanquartier, Fleur, Claire Jean-Quartier, Sarah Stryeck, and Andreas Holzinger. 2021. "Open Data to Support CANCER Science—A Bioinformatics Perspective on Glioma Research" Onco 1, no. 2: 219-229. https://doi.org/10.3390/onco1020016
APA StyleJeanquartier, F., Jean-Quartier, C., Stryeck, S., & Holzinger, A. (2021). Open Data to Support CANCER Science—A Bioinformatics Perspective on Glioma Research. Onco, 1(2), 219-229. https://doi.org/10.3390/onco1020016