The Effects of Particle LET and Fluence on the Complexity and Frequency of Clustered DNA Damage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Radiation Transport Calculation
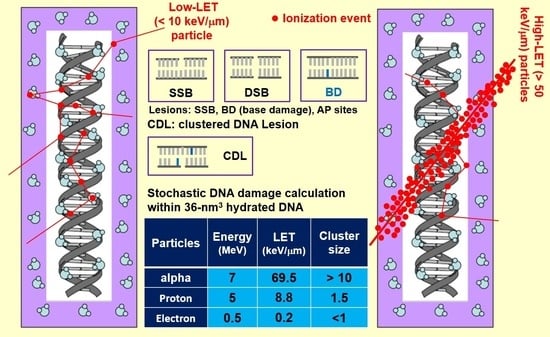
2.2. DNA Damage Calculation
2.3. Statistical Uncertainty
3. Results
3.1. Ionization Density
3.2. Characterization of LEEs
3.3. Reactive Species from Water Radiolysis
3.4. Quantification of DNA Damage
3.5. Cluster Size and Damage Site
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paganetti, H. Relative biological effectiveness (RBE) values for proton beam therapy. Variations as a function of biological endpoint, dose, and linear energy transfer. Phys. Med. Biol. 2014, 59, 419. [Google Scholar] [CrossRef] [PubMed]
- Goodhead, D.T. Initial events in the cellular effects of ionizing radiations: Clustered damage in DNA. Int. J. Radiat. Biol. 1994, 65, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Blakely, E.A. The 20th gray lecture 2019: Health and heavy ions. Br. J. Radiol. 2020, 93, 20200172. [Google Scholar] [CrossRef] [PubMed]
- Goodhead, D.T. Molecular and cell models of biological effects of heavy ion radiation. Radiat. Environ. Biophys. 1995, 34, 67–72. [Google Scholar] [CrossRef]
- Nikjoo, H.; Emfietzoglou, D.; Liamsuwan, T.; Taleei, R.; Liljequist, D.; Uehara, S. Radiation track, DNA damage and response—A review. Rep. Prog. Phys. 2016, 79, 116601. [Google Scholar] [CrossRef]
- Lomax, M.E.; Folkes, L.K.; O’Neill, P. Biological consequences of radiation-induced DNA damage: Relevance to radiotherapy. Clin. Oncol. 2013, 25, 578–585. [Google Scholar] [CrossRef]
- Johnston, P.J.; Bryant, P.E. A component of DNA double-strand break repair is dependent on the spatial orientation of the lesions within the higher-order structures of chromatin. Int. J. Radiat. Biol. 1994, 66, 531–536. [Google Scholar] [CrossRef]
- Eccles, L.J.; O’Neill, P.; Lomax, M.E. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res./Fundam. Mol. Mech. Mutagen. 2011, 711, 134–141. [Google Scholar] [CrossRef]
- Lorat, Y.; Brunner, C.U.; Schanz, S.; Jakob, B.; Taucher-Scholz, G.; Rübe, C.E. Nanoscale analysis of clustered DNA damage after high-LET irradiation by quantitative electron microscopy—The heavy burden to repair. DNA Repair. 2015, 28, 93–106. [Google Scholar] [CrossRef]
- Hada, M.; Georgakilas, A.G. Formation of clustered DNA damage after high-LET irradiation: A review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef]
- Adhikary, A.; Becker, D.; Sevilla, M.D. Electron spin resonance of radicals in irradiated DNA. In Applications of EPR in Radiation Research; Lund, A., Shiotani, M., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 299–352. [Google Scholar] [CrossRef]
- Nakano, T.; Akamatsu, K.; Tsuda, M.; Tujimoto, A.; Hirayama, R.; Hiromoto, T.; Tamada, T.; Ide, H.; Shikazono, N. Formation of clustered DNA damage in vivo upon irradiation with ionizing radiation: Visualization and analysis with atomic force microscopy. Proc. Natl. Acad. Sci. USA 2022, 119, e2119132119. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Nakano, T.; Tsuda, M.; Kanamoto, R.; Hirayama, R.; Uzawa, A.; Ide, H. Direct observation of damage clustering in irradiated DNA with atomic force microscopy. Nucleic Acids Res. 2020, 48, e18. [Google Scholar] [CrossRef] [PubMed]
- Gulston, M.; de Lara, C.; Jenner, T.; Davis, E.; O’Neill, P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004, 32, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Chatzipapas, K.P.; Papadimitroulas, P.; Emfietzoglou, D.; Kalospyros, S.A.; Hada, M.; Georgakilas, A.G.; Kagadis, G.C. Ionizing radiation and complex DNA damage: Quantifying the radiobiological damage using monte carlo simulations. Cancers 2020, 12, 799. [Google Scholar] [CrossRef] [PubMed]
- Bertolet, A.; Ramos-Méndez, J.; McNamara, A.; Yoo, D.; Ingram, S.; Henthorn, N.; Warmenhoven, J.-W.; Faddegon, B.; Merchant, M.; McMahon, S.J.; et al. Impact of DNA geometry and scoring on monte carlo track-structure simulations of initial radiation-induced damage. Radiat. Res. 2022, 198, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Sakata, D.; Belov, O.; Bordage, M.; Emfietzoglou, D.; Guatelli, S.; Inaniwa, T.; Ivanchenko, V.; Karamitros, M.; Kyriakou, I.; Lampe, N.; et al. Fully integrated monte carlo simulation for evaluating radiation induced DNA damage and subsequent repair using Geant4-DNA. Sci. Rep. 2020, 10, 20788. [Google Scholar] [CrossRef]
- Alizadeh, E.; Orlando, T.M.; Sanche, L. Biomolecular damage induced by ionizing radiation: The direct and indirect effects of low-energy electrons on DNA. Annu. Rev. Phys. Chem. 2015, 66, 379–398. [Google Scholar] [CrossRef]
- Becker, D.; Kumar, A.; Adhikary, A.; Sevilla, M.D. Gamma- and ion-beam DNA radiation damage: Theory and experiment. In DNA Damage, DNA Repair and Disease: Volume 2; Dizdaroglu, M., Lloyd, R.S., Dizdaroglu, M., Lloyd, R.S., Eds.; The Royal Society of Chemistry: London, UK, 2020; pp. 426–457. [Google Scholar] [CrossRef]
- Muraro, S.; Battistoni, G.; Kraan, A.C. Challenges in monte carlo simulations as clinical and research tool in particle therapy: A review. Front. Phys. 2020, 8, 567800. [Google Scholar] [CrossRef]
- Baba, K.; Kusumoto, T.; Okada, S.; Ogawara, R.; Kodaira, S.; Raffy, Q.; Barillon, R.; Ludwig, N.; Galindo, C.; Peaupardin, P.; et al. Quantitative estimation of track segment yields of water radiolysis species under heavy ions around Bragg peak energies using Geant4-DNA. Sci. Rep. 2021, 11, 1524. [Google Scholar] [CrossRef]
- Nikjoo, H.; O’Neill, P.; Wilson, W.E.; Goodhead, D.T. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 2001, 156, 577–583. [Google Scholar] [CrossRef]
- Sanche, L. Interaction of low energy electrons with DNA: Applications to cancer radiation therapy. Radiat. Phys. Chem. 2016, 128, 36–43. [Google Scholar] [CrossRef]
- Pimblott, S.M.; LaVerne, J.A. Production of low-energy electrons by ionizing radiation. Radiat. Phys. Chem. 2007, 76, 1244–1247. [Google Scholar] [CrossRef]
- Narayanan, S.J.J.; Tripathi, D.; Verma, P.; Adhikary, A.; Dutta, A.K. Secondary electron attachment-induced radiation damage to genetic materials. ACS Omega 2023, 8, 10669–10689. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Kumar, A.; Muroya, Y.; Yamashita, S.; Sakurai, T.; Denisov, S.A.; Sevilla, M.D.; Adhikary, A.; Seki, S.; Mostafavi, M. Observation of dissociative quasi-free electron attachment to nucleoside via excited anion radical in solution. Nat. Commun. 2019, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Jiang, S.B.; Jia, X. Accelerated monte carlo simulation on the chemical stage in water radiolysis using GPU. Phys. Med. Biol. 2017, 62, 3081. [Google Scholar] [CrossRef]
- Friedland, W.; Dingfelder, M.; Kundrát, P.; Jacob, P. Track structures, DNA targets and radiation effects in the biophysical monte carlo simulation code PARTRAC. Mutat. Res. 2011, 711, 28–40. [Google Scholar] [CrossRef]
- Halliwell, B.; Adhikary, A.; Dingfelder, M.; Dizdaroglu, M. Hydroxyl radical is a significant player in oxidative DNA damage in vivo. Chem. Soc. Rev. 2021, 50, 8355–8360. [Google Scholar] [CrossRef]
- von Sonntag, C. Free-Radical-Induced DNA Damage and Its Repair: A Chemical Perspective; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Milano, M.T.; Bernhard, W.A. The effect of packing and conformation on free radical yields in films of variably hydrated DNA. Radiat. Res. 1999, 151, 39–49. [Google Scholar] [CrossRef]
- Purkayastha, S.; Milligan, J.R.; Bernhard, W.A. The role of hydration in the distribution of free radical trapping in directly ionized DNA. Radiat. Res. 2006, 166, 1–8. [Google Scholar] [CrossRef]
- Kyriaknu, I.; Sakata, D.; Tran, H.N.; Perrot, Y.; Shin, W.G.; Lampe, N.; Zein, S.A.; Bordage, M.; Guatelli, S.; Villagrasa, C.; et al. Review of the GEANT4-DNA simulation toolkit for radiobiological applications at the cellular and DNA level. Cancers 2022, 14, 35. [Google Scholar] [CrossRef]
- Adjei, D.; Trinh, N.D.; Mostafavi, M. Application of Geant4-DNA for simulating water radiolysis induced by auger electron-emitting radionuclides. J. Radiat. Res. 2023, 64, 369–378. [Google Scholar] [CrossRef]
- Villagrasa, C.; Francis, Z.; Incerti, S. Physical models implemented in the GEANT4-DNA extension of the GEANT-4 toolkit for calculating initial radiation damage at the molecular level. Radiat. Prot. Dosim. 2011, 143, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Zein, S.A.; Bordage, M.; Tran, H.N.; Macetti, G.; Genoni, A.; Cappello, C.D.; Incerti, S. Monte carlo simulations of electron interactions with the DNA molecule: A complete set of physics models for Geant4-DNA simulation toolkit. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2023, 542, 51–60. [Google Scholar] [CrossRef]
- Champion, C.; Incerti, S.; Tran, H.N.; Karamitros, M.; Shin, J.; Lee, S.; Lekadir, H.; Bernal, M.; Francis, Z.; Ivanchenko, V. Proton transport in water and DNA components: A Geant4 monte carlo simulation. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2013, 306, 165–168. [Google Scholar] [CrossRef]
- Incerti, S.; Kyriakou, I.; Bernal, M.A.; Bordage, M.C.; Francis, Z.; Guatelli, S.; Ivanchenko, V.; Karamitros, M.; Lampe, N.; Lee, S.B. Geant4-DNA example applications for track structure simulations in liquid water: A report from the Geant4-DNA project. Med. Phys. 2018, 45, e722–e739. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R. (Ed.) The Nucleus—Volume 1: Nuclei and Subnuclear Components; Methods in Molecular Biology 463; Humana Press: Totowa, NJ, USA, 2008. [Google Scholar]
- Khanduri, D.; Adhikary, A.; Sevilla, M.D. Highly Oxidizing Excited States of One-Electron Oxidized Guanine in DNA: Wavelength and pH Dependence. J. Am. Chem. Soc. 2011, 133, 4527–4537. [Google Scholar] [CrossRef] [PubMed]
- Śmiałek, M.A.; Jones, N.C.; Hoffmann, S.V.; Mason, N.J. Measuring the density of DNA films using ultraviolet-visible interferometry. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2013, 87, 060701. [Google Scholar] [CrossRef]
- Fasman, G.D. Handbook of Biochemistry and Molecular Biology, 3rd ed.; CRC Press: Cleveland, OH, USA, 1977. [Google Scholar]
- Rezaee, M.; Cloutier, P.; Bass, A.D.; Michaud, M.; Hunting, D.J.; Sanche, L. Absolute cross section for low-energy-electron damage to condensed macromolecules: A case study of DNA. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2012, 86, 031913. [Google Scholar] [CrossRef]
- Kouass Sahbani, S.; Sanche, L.; Cloutier, P.; Bass, A.D.; Hunting, D.J. Loss of cellular transformation efficiency induced by DNA irradiation with low-energy (10 eV) electrons. J. Phys. Chem. B 2014, 118, 13123–13131. [Google Scholar] [CrossRef]
- Ito, T.; Baker, S.; Stickley, C.; Peak, J.; Peak, M. Dependence of the Yield of Strand Breaks Induced by γ-rays in DNA on the Physical Conditions of Exposure: Water Content and Temperature. Int. J. Radiat. Biol. 1993, 63, 289–296. [Google Scholar] [CrossRef]
- Greenberg, M.D. Radical and Radical Ion Reactivity in Nucleic Acid Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Sevilla, M.D.; Becker, D.; Kumar, A.; Adhikary, A. Radiation-Induced Free Radical Mechanisms in DNA. Radiat. Phys. Chem. 2016, 128, 60–74. [Google Scholar] [CrossRef]
- Siddiqi, M.A.; Bothe, E. Single- and Double-Strand Break Formation in DNA Irradiated in Aqueous Solution: Dependence on Dose and OH Radical Scavenger Concentration. Radiat. Res. 1987, 112, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Isabelle, V.; Prévost, C.; Spotheim-Maurizot, M.; Sabattier, R.; Charlier, M. Radiation-induced Damages in Single- and Double-stranded DNA. Int. J. Radiat. Biol. 1995, 67, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.; Adhikary, A.; Tetteh, S.T.; Bull, A.W.; Sevilla, M.D. Kr-86 Ion-Beam Irradiation of Hydrated DNA: Free Radical and Unaltered Base Yields. Radiat. Res. 2012, 178, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Kant, M.; Jaruga, P.; Coskun, E.; Ward, S.; Stark, A.D.; Baumann, T.; Becker, D.; Adhikary, A.; Sevilla, M.D.; Dizdaroglu, M. Ne-22 Ion-Beam Radiation Damage to DNA: From Initial Free Radical Formation to Resulting DNA-Base Damage. ACS Omega 2021, 6, 16600–16611. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Ravanat, J.-L.; TavernaPorro, M.; Menoni, H.; Angelov, D. Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett. 2012, 327, 5–15. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Coskun, E.; Jaruga, P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic. Res. 2015, 49, 525–548. [Google Scholar] [CrossRef]
- Kim, H.-K.; Titze, J.; Schöffler, M.; Trinter, F.; Waitz, M.; Voigtsberger, J.; Sann, H.; Meckel, M.; Stuck, C.; Lenz, U.; et al. Enhanced production of low energy electrons by alpha particle impact. Proc. Natl. Acad. Sci. USA. 2011, 108, 11821–11824. [Google Scholar] [CrossRef]
- LaVerne, J.A. The Production of OH Radicals in the Radiolysis of Water with 4He Ions. Radiat. Res. 1989, 118, 201–210. [Google Scholar] [CrossRef]
- Bertolet, A.; Ramos-Méndez, J.; Paganetti, H.; Schuemann, J. The relation between microdosimetry and induction of direct damage to DNA by alpha particles. Phys. Med. Biol. 2021, 66, 155016. [Google Scholar] [CrossRef]
- Gurzadyan, G.G.; Görner, H. Photolesions in DNA upon 193 nm excitation. Photochem. Photobiol. 1993, 58, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Bowman, M.K.; Becker, D.; Sevilla, M.D.; Zimbrick, J.D. Track Structure in DNA Irradiated with Heavy Ions. Radiat. Res. 2005, 163, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Wardman, P. Radiotherapy using high-intensity pulsed radiation beams (FLASH): A radiation-chemical perspective. Radiat. Res. 2020, 194, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Jonah, C.D.; Hart, E.J.; Matheson, M.S. Yields and Decay of the Hydrated Electron at Times Greater Than 200 Picoseconds. J. Phys. Chem. 1973, 77, 1838–1843. [Google Scholar] [CrossRef]
- Favaudon, V.; Caplier, L.; Monceau, V.; Pouzoulet, F.; Sayarath, M.; Fouillade, C.; Poupon, M.F.; Brito, I.; Hupe, P.H.; Hall, J.; et al. Ultrahigh dose-rate FLASH irradiation increases the differential response between normal and tumor tissue in mice. Sci. Transl. Med. 2014, 6, 245ra93. [Google Scholar] [CrossRef] [PubMed]
- Guan, F.; Peeler, C.; Bronk, L.; Geng, C.; Taleei, R.; Randeniya, S.; Ge, S.; Mirkovic, D.; Grosshans, D.; Mohan, R.; et al. Analysis of the track- and dose-averaged LET and LET spectra in proton therapy using the geant4 Monte Carlo code. Med. Phys. 2015, 42, 6234–6247. [Google Scholar] [CrossRef] [PubMed]
- Liberal, F.D.C.G.; Moreira, H.; Redmond, K.M.; O’sullivan, J.M.; Alshehri, A.H.D.; Wright, T.C.; Dunne, V.L.; Campfield, C.; Biggart, S.; McMahon, S.J.; et al. Differential responses to 223Ra and Alpha-particles exposure in prostate cancer driven by mitotic catastrophe. Front. Oncol. 2022, 12, 877302. [Google Scholar] [CrossRef] [PubMed]
- Penninckx, S.; Cekanaviciute, E.; Degorre, C.; Guiet, E.; Viger, L.; Lucas, S.; Costes, S.V. Dose, LET and Strain Dependence of Radiation-Induced 53BP1 Foci in 15 Mouse Strains Ex Vivo Introducing Novel DNA Damage Metrics. Radiat. Res. 2019, 192, 1–12. [Google Scholar] [CrossRef]
- Particle Therapy Co-Operative Group (PTCOG). Available online: https://www.ptcog.site/index.php/facilities-in-operation-public (accessed on 16 December 2023).
- James, S.S.; Bednarz, B.; Benedict, S.; Buchsbaum, J.C.; Dewaraja, Y.; Frey, E.; Hobbs, R.; Grudzinski, J.; Roncali, E.; Sgouros, G.; et al. Current Status of Radiopharmaceutical Therapy. Int. J. Radiat. Oncol. 2021, 109, 891–901. [Google Scholar] [CrossRef]
- Paul, J.M.; Koch, R.F.; Philip, P.C. AAPM Task Group 21 protocol: Dosimetric evaluation. Med. Phys. 1985, 12, 424–430. [Google Scholar] [CrossRef]
- Almond, P.R.; Biggs, P.J.; Coursey, B.M.; Hanson, W.F.; Huq, M.S.; Nath, R.; Rogers, D.W.O. AAPM’s TG-51 protocol for clinical reference dosimetry of high-energy photon and electron beams. Med. Phys. 1999, 26, 1847–1870. [Google Scholar] [CrossRef] [PubMed]
- Kalholm, F.; Grzanka, L.; Traneus, E.; Bassler, N. A systematic review on the usage of averaged LET in radiation biology for particle therapy. Radiother. Oncol. 2021, 161, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Danforth, J.M.; Provencher, L.; Goodarzi, A.A. Chromatin and the Cellular Response to Particle Radiation-Induced Oxidative and Clustered DNA Damage. Front. Cell Dev. Biol. 2022, 10, 910440. [Google Scholar] [CrossRef] [PubMed]
- Prise, K. Use of Radiation Quality as a Probe for DNA Lesion Complexity. Int. J. Radiat. Biol. 1994, 65, 43–48. [Google Scholar] [CrossRef]
- Rezaee, M.; Hill, R.P.; Jaffray, D.A. The Exploitation of Low-Energy Electrons in Cancer Treatment. Radiat. Res. 2017, 188, 123–143. [Google Scholar] [CrossRef] [PubMed]
- LaVerne, J.A.; Pimblott, S.M. Electron Energy-Loss Distributions in Solid, Dry DNA. Radiat. Res. 1995, 141, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Sahbani, S.K.; Girouard, S.; Cloutier, P.; Sanche, L.; Hunting, D.J. The Relative Contributions of DNA Strand Breaks, Base Damage and Clustered Lesions to the Loss of DNA Functionality Induced by Ionizing Radiation. Radiat. Res. 2014, 181, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zheng, Y.; Sanche, L. DNA strand breaks and crosslinks induced by transient anions in the range 2–20 eV. J. Chem. Phys. 2014, 140, 155101. [Google Scholar] [CrossRef]
- Wang, X.; Liao, H.; Liu, W.; Shao, Y.; Zheng, Y.; Sanche, L. DNA Protection against Damages Induced by Low-Energy Electrons: Absolute Cross Sections for Arginine–DNA Complexes. J. Phys. Chem. Lett. 2023, 14, 5674–5680. [Google Scholar] [CrossRef]
- Lemelin, V.; Sanche, L. High-resolution electron energy loss spectroscopy: Absolute cross section measurements for low energy electron scattering from biomolecules. In Radiation in Bioanalysis: Spectroscopic Techniques and Theoretical Methods; Pereira, A.S., Tavares, P., Limão-Vieira, P., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 3–42. [Google Scholar] [CrossRef]
- Zheng, Y.; Sanche, L. Effective and absolute cross sections for low-energy (1–30 eV) electron interactions with condensed biomolecules. Appl. Phys. Rev. 2018, 5, 021302. [Google Scholar] [CrossRef]
- Yamashita, S.; Taguchi, M.; Baldacchino, G.; Katsumura, Y. Radiation chemistry of liquid water with heavy ions: Steady-state and pulse radiolysis studies. In Charged Particle and Photon Interactions with Matter; Hatano, Y., Katsumura, Y., Mozumder, A., Eds.; Taylor and Francis Group: Boca Raton, FL, USA, 2010. [Google Scholar]
- Smith, M.; Pimblott, S.M.; LaVerne, J.A. Hydroxyl radical yields in the heavy ion radiolysis of water. Radiat. Phys. Chem. 2021, 188, 109629. [Google Scholar] [CrossRef]
- El Omar, A.K.; Schmidhammer, U.; Jeunesse, P.; Larbre, J.-P.; Lin, M.; Muroya, Y.; Katsumura, Y.; Pernot, P.; Mostafavi, M. Time-Dependent Radiolytic Yield of OH• Radical Studied by Picosecond Pulse Radiolysis. J. Phys. Chem. A 2011, 115, 12212–12216. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, M.; Kimura, A.; Watanabe, R.; Hirota, K. Estimation of Yields of Hydroxyl Radicals in Water under Various Energy Heavy Ions. Radiat. Res. 2009, 171, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Pastina, B.; LaVerne, J.A. Hydrogen Peroxide Production in the Radiolysis of Water with Heavy Ions. J. Phys. Chem. A 1999, 103, 1592–1597. [Google Scholar] [CrossRef]
- Wasselin-Trupin, V.; Baldacchino, G.; Bouffard, S.; Hickel, B. Hydrogen peroxide yields in water radiolysis by high-energy ion beams at constant LET. Radiat. Phys. Chem. 2002, 65, 53–61. [Google Scholar] [CrossRef]
- Krisch, R.E.; Flick, M.B.; Trumbore, C.N. Radiation Chemical Mechanisms of Single- and Double-Strand Break Formation in Irradiated SV40 DNA. Radiat. Res. 1991, 126, 251–259. [Google Scholar] [CrossRef]
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef]






| Alpha | Proton | Electron | ||||||
|---|---|---|---|---|---|---|---|---|
| Energy (MeV) | Initial LET (keV/μm) | Track-Averaged LET (keV/μm) | Energy (MeV) | Initial LET (keV/μm) | Track-Averaged LET (keV/μm) | Energy (keV) | Initial LET (keV/μm) | Track-Averaged LET (keV/μm) |
| 0.75 | 233.8 | 262.2 | 0.1 | 70.6 | 81.5 | 0.1 | 28.7 | 28.7 |
| 1 | 219 | 221.7 | 0.2 | 66.0 | 66.2 | 0.2 | 26.6 | 28.5 |
| 2 | 161.4 | 162.4 | 0.5 | 41.0 | 41.3 | 0.5 | 17 | 18.6 |
| 4 | 101.3 | 103.4 | 1 | 26.7 | 28.0 | 1 | 11.2 | 12.5 |
| 7 | 67.4 | 69.5 | 2 | 15.9 | 16.9 | 2 | 7.1 | 7.3 |
| 10 | 51.5 | 53.4 | 5 | 7.9 | 8.8 | 5 | 3.6 | 3.8 |
| 20 | 30.6 | 31.4 | 10 | 4.8 | 5.0 | 10 | 2.1 | 2.2 |
| 50 | 14.6 | 15.2 | 20 | 2.6 | 2.8 | 50 | 0.7 | 0.7 |
| 100 | 8.1 | 8.6 | 50 | 1.2 | 1.3 | 100 | 0.4 | 0.4 |
| 200 | 4.5 | 4.9 | 100 | 0.7 | 0.7 | 500 | 0.2 | 0.2 |
| Particles | Low Energy, High LET Particles | High Energy, Medium- and Low-LET Particles | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Energy (MeV) | LET (keV/μm) | Range (keV/μm) | Cluster Size per Voxel Volume | Number of Damage Site | Energy (MeV) | LET (keV/μm) | Range (keV/μm) | Cluster Size per Voxel Volume | Number of Damage Site | |
| α | 2 | 162.4 | 11.2 | 39.8 | 1.6 | 50 | 15.2 | 1.8 | 2.8 | 17.3 |
| 4 | 103.4 | 27.1 | 23.1 | 2.5 | 100 | 8.6 | 6.4 | 1.4 | 30.3 | |
| p | 0.5 | 41.3 | 8.9 | 10.8 | 6.3 | 50 | 1.2 | 22.3 | <1 | 47.4 |
| 1 | 26.1 | 24.6 | 6.3 | 10.1 | 100 | 0.7 | 77.2 | <1 | 47.1 | |
| e | NA | NA | NA | NA | NA | 0.1 | 0.4 | 0.14 | <1 | 67.9 |
| 0.5 | 0.2 | 1.8 | <1 | 86.7 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezaee, M.; Adhikary, A. The Effects of Particle LET and Fluence on the Complexity and Frequency of Clustered DNA Damage. DNA 2024, 4, 34-51. https://doi.org/10.3390/dna4010002
Rezaee M, Adhikary A. The Effects of Particle LET and Fluence on the Complexity and Frequency of Clustered DNA Damage. DNA. 2024; 4(1):34-51. https://doi.org/10.3390/dna4010002
Chicago/Turabian StyleRezaee, Mohammad, and Amitava Adhikary. 2024. "The Effects of Particle LET and Fluence on the Complexity and Frequency of Clustered DNA Damage" DNA 4, no. 1: 34-51. https://doi.org/10.3390/dna4010002
APA StyleRezaee, M., & Adhikary, A. (2024). The Effects of Particle LET and Fluence on the Complexity and Frequency of Clustered DNA Damage. DNA, 4(1), 34-51. https://doi.org/10.3390/dna4010002