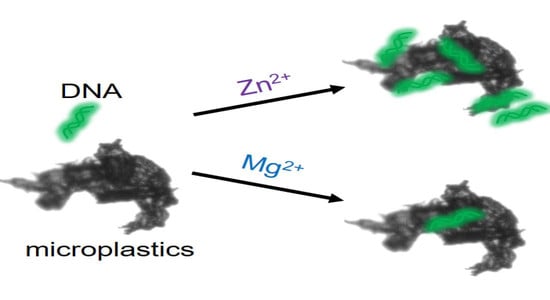
Promotion of DNA Adsorption onto Microplastics by Transition Metal Ions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Microplastics
2.3. Purification of Salmon Sperm DNA
2.4. Adsorption of ssDNA and Salmon dsDNA
2.5. Adsorption of Short dsDNA
2.6. Desorption of DNA from Microplastics
3. Results and Discussion
3.1. Preparation and Characterization of the Microplastics
3.2. Comparison of Mg2+, Mn2+ and Zn2+ for ssDNA Adsorption
3.3. Adsorption of dsDNA
3.4. Desorption of dsDNA to Probe Adsorption Affinity
3.5. Testing DNA Adsorption in Wastewater
3.6. Additional Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, Use, and Fate of All Plastics Ever Made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLeod, M.; Arp, H.P.H.; Tekman, M.B.; Jahnke, A. The Global Threat from Plastic Pollution. Science 2021, 373, 61–65. [Google Scholar] [CrossRef]
- Ivleva, N.P. Chemical Analysis of Microplastics and Nanoplastics: Challenges, Advanced Methods, and Perspectives. Chem. Rev. 2021, 121, 11886–11936. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, J.; Zhang, W.; Shi, L.; Yi, K.; Yu, H.; Zhang, C.; Li, S.; Li, J. Microplastics as a Vehicle of Heavy Metals in Aquatic Environments: A Review of Adsorption Factors, Mechanisms, and Biological Effects. J. Environ. Manag. 2022, 302, 113995. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, T.T.S.; Andreu, I.; Machado, M.C.; Vimbela, G.; Tripathi, A.; Bose, A. Interaction of Cyanobacteria with Nanometer and Micron Sized Polystyrene Particles in Marine and Fresh Water. Langmuir 2020, 36, 3963–3969. [Google Scholar] [CrossRef]
- Ju, S.; Shin, G.; Lee, M.; Koo, J.M.; Jeon, H.; Ok, Y.S.; Hwang, D.S.; Hwang, S.Y.; Oh, D.X.; Park, J. Biodegradable Chito-Beads Replacing Non-Biodegradable Microplastics for Cosmetics. Green Chem. 2021, 23, 6953–6965. [Google Scholar] [CrossRef]
- Wright, S.L.; Kelly, F.J. Plastic and Human Health: A Micro Issue? Environ. Sci. Technol. 2017, 51, 6634–6647. [Google Scholar] [CrossRef]
- Cox, K.D.; Covernton, G.A.; Davies, H.L.; Dower, J.F.; Juanes, F.; Dudas, S.E. Human Consumption of Microplastics. Environ. Sci. Technol. 2019, 53, 7068–7074. [Google Scholar] [CrossRef] [Green Version]
- Hildebrandt, L.; Nack, F.; Zimmermann, T.; Pröfrock, D. Microplastics as a Trojan Horse for Trace Metals. J. Hazard. Mater. Lett. 2021, 2, 100035. [Google Scholar] [CrossRef]
- Lee, A.; Mondon, J.; Merenda, A.; Dumée, L.F.; Callahan, D.L. Surface Adsorption of Metallic Species onto Microplastics with Long-Term Exposure to the Natural Marine Environment. Sci. Total Environ. 2021, 780, 146613. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, X.; Tam, N.F.; Lao, X.; Zhong, M.; Wu, Q.; Lei, H.; Chen, Z.; Li, Z.; Fu, J. An Insight into Aggregation Kinetics of Polystyrene Nanoplastics Interaction with Metal Cations. Chin. Chem. Lett. 2022, 33, 5213–5217. [Google Scholar] [CrossRef]
- Cao, Y.; Zhao, M.; Ma, X.; Song, Y.; Zuo, S.; Li, H.; Deng, W. A Critical Review on the Interactions of Microplastics with Heavy Metals: Mechanism and Their Combined Effect on Organisms and Humans. Sci. Total Environ. 2021, 788, 147620. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Saran, R.; Liu, J. Metal Sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Xie, Y.; Li, Y.; Huang, Y.; Parent, L.R.; Ditri, T.; Zang, N.; Rinehart, J.D.; Gianneschi, N.C. Tunable, Metal-Loaded Polydopamine Nanoparticles Analyzed by Magnetometry. Chem. Mater. 2017, 29, 8195–8201. [Google Scholar] [CrossRef]
- Valencia, L.; Monti, S.; Kumar, S.; Zhu, C.; Liu, P.; Yu, S.; Mathew, A.P. Nanocellulose/Graphene Oxide Layered Membranes: Elucidating Their Behaviour During Filtration of Water and Metal Ions in Real Time. Nanoscale 2019, 11, 22413–22422. [Google Scholar] [CrossRef] [Green Version]
- Kushalkar, M.P.; Liu, B.; Liu, J. Promoting DNA Adsorption by Acids and Polyvalent Cations: Beyond Charge Screening. Langmuir 2020, 36, 11183–11195. [Google Scholar] [CrossRef]
- Zandieh, M.; Liu, J. Metal-Doped Polydopamine Nanoparticles for Highly Robust and Efficient DNA Adsorption and Sensing. Langmuir 2021, 37, 8953–8960. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, Z.; Han, J.; Xie, G.; Liu, J. Polyvalent Metal Ion Promoted Adsorption of DNA Oligonucleotides by Montmorillonite. Langmuir 2021, 37, 1037–1044. [Google Scholar] [CrossRef]
- Zandieh, M.; Patel, K.; Liu, J. Adsorption of Linear and Spherical DNA Oligonucleotides onto Microplastics. Langmuir 2022, 38, 1915–1922. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of Aptamer Discovery and Technology. Nat. Rev. Chem. 2017, 1, 76. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, Z.; Yang, R.; Liu, J. Selection and Screening of DNA Aptamers for Inorganic Nanomaterials. Chem. Eur. J. 2018, 24, 2525–2532. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. 2021, 60, 16800–16823. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, H.; Qi, Z.; Huang, Z.; Ma, L.; Liu, J. Freezing-Assisted Conjugation of Unmodified Diblock DNA to Hydrogel Nanoparticles and Monoliths for DNA and Hg2+ Sensing. Angew. Chem. Int. Ed. 2021, 60, 12985–12991. [Google Scholar] [CrossRef] [PubMed]
- Zandieh, M.; Liu, J. Removal and Degradation of Microplastics Using the Magnetic and Nanozyme Activities of Bare Iron Oxide Nanoaggregates. Angew. Chem. Int. Ed. 2022, 61, e202212013. [Google Scholar] [CrossRef]
- Hulme, E.C.; Trevethick, M.A. Ligand Binding Assays at Equilibrium: Validation and Interpretation. Br. J. Pharmacol. 2010, 161, 1219–1237. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Mou, H.; Zhao, X.; Wang, Y.; Mu, T. Eutectic Molecular Liquids Based on Hydrogen Bonding and π–π Interaction for Exfoliating Two-Dimensional Materials and Recycling Polymers. Chem. Asian J. 2019, 14, 3350–3356. [Google Scholar] [CrossRef]
- Ni, Y.-P.; Li, Q.-T.; Chen, L.; Wu, W.-S.; Qin, Z.-H.; Zhang, Y.; Chen, L.; Wang, X.-L.; Wang, Y.-Z. Semi-Aromatic Copolyesters with High Strength and Fire Safety Via Hydrogen Bonds and Π-Π Stacking. Chem. Eng. J. 2019, 374, 694–705. [Google Scholar] [CrossRef]
- Zandieh, M.; Liu, J. Transition Metal-Mediated DNA Adsorption on Polydopamine Nanoparticles. Langmuir 2020, 36, 3260–3267. [Google Scholar] [CrossRef]
- Zandieh, M.; Liu, J. Cooperative Metal Ion-Mediated Adsorption of Spherical Nucleic Acids with a Large Hysteresis. Langmuir 2020, 36, 14324–14332. [Google Scholar] [CrossRef]
- Dong, H.; Chen, Y.; Wang, J.; Zhang, Y.; Zhang, P.; Li, X.; Zou, J.; Zhou, A. Interactions of Microplastics and Antibiotic Resistance Genes and Their Effects on the Aquaculture Environments. J. Hazard. Mater. 2021, 403, 123961. [Google Scholar] [CrossRef]
- Yuan, Q.; Sun, R.; Yu, P.; Cheng, Y.; Wu, W.; Bao, J.; Alvarez, P.J. UV-Aging of Microplastics Increases Proximal ARG Donor-Recipient Adsorption and Leaching of Chemicals That Synergistically Enhance Antibiotic Resistance Propagation. J. Hazard. Mater. 2022, 427, 127895. [Google Scholar] [CrossRef]
- Glasel, J. Validity of Nucleic Acid Purities Monitored by 260nm/280nm Absorbance Ratios. BioTechniques 1995, 18, 62–63. [Google Scholar]
- Lucena-Aguilar, G.; Sánchez-López, A.M.; Barberán-Aceituno, C.; Carrillo-Avila, J.A.; López-Guerrero, J.A.; Aguilar-Quesada, R. DNA Source Selection for Downstream Applications Based on DNA Quality Indicators Analysis. Biopreserv. Biobank. 2016, 14, 264–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Sun, Z.; Zhang, X.; Liu, J. Mechanisms of DNA Sensing on Graphene Oxide. Anal. Chem. 2013, 85, 7987–7993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mammen, M.; Choi, S.K.; Whitesides, G.M. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem. Int. Ed. 1998, 37, 2754–2794. [Google Scholar] [CrossRef]
- Lu, C.-H.; Zhu, C.-L.; Li, J.; Liu, J.-J.; Chen, X.; Yang, H.-H. Using Graphene to Protect DNA from Cleavage During Cellular Delivery. Chem. Commun. 2010, 46, 3116–3118. [Google Scholar] [CrossRef] [Green Version]
- Rollion-Bard, C.; Saulnier, S.; Vigier, N.; Schumacher, A.; Chaussidon, M.; Lécuyer, C. Variability in Magnesium, Carbon and Oxygen Isotope Compositions of Brachiopod Shells: Implications for Paleoceanographic Studies. Chem. Geol. 2016, 423, 49–60. [Google Scholar] [CrossRef]
- Andersen, M.B.; Vance, D.; Archer, C.; Anderson, R.F.; Ellwood, M.J.; Allen, C.S. The Zn Abundance and Isotopic Composition of Diatom Frustules, a Proxy for Zn Availability in Ocean Surface Seawater. Earth Planet. Sci. Lett. 2011, 301, 137–145. [Google Scholar] [CrossRef]
- Liu, B.; Ma, L.; Huang, Z.; Hu, H.; Wu, P.; Liu, J. Janus DNA Orthogonal Adsorption of Graphene Oxide and Metal Oxide Nanoparticles Enabling Stable Sensing in Serum. Mater. Horiz. 2018, 5, 65–69. [Google Scholar] [CrossRef]
- Tong, F.; Liu, D.; Zhang, Z.; Chen, W.; Fan, G.; Gao, Y.; Gu, X.; Gu, C. Heavy Metal-Mediated Adsorption of Antibiotic Tetracycline and Ciprofloxacin on Two Microplastics: Insights into the Role of Complexation. Environ. Res. 2023, 216, 114716. [Google Scholar] [CrossRef]
- Hao, J.; Mokhtari, M.; Pedreira-Segade, U.; Michot, L.J.; Daniel, I. Transition Metals Enhance the Adsorption of Nucleotides onto Clays: Implications for the Origin of Life. ACS Earth Space Chem. 2018, 3, 109–119. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Patel, K.; Zandieh, M.; Liu, J. Promotion of DNA Adsorption onto Microplastics by Transition Metal Ions. Microplastics 2023, 2, 158-167. https://doi.org/10.3390/microplastics2010012
Wu L, Patel K, Zandieh M, Liu J. Promotion of DNA Adsorption onto Microplastics by Transition Metal Ions. Microplastics. 2023; 2(1):158-167. https://doi.org/10.3390/microplastics2010012
Chicago/Turabian StyleWu, Lyuyuan, Kshiti Patel, Mohamad Zandieh, and Juewen Liu. 2023. "Promotion of DNA Adsorption onto Microplastics by Transition Metal Ions" Microplastics 2, no. 1: 158-167. https://doi.org/10.3390/microplastics2010012
APA StyleWu, L., Patel, K., Zandieh, M., & Liu, J. (2023). Promotion of DNA Adsorption onto Microplastics by Transition Metal Ions. Microplastics, 2(1), 158-167. https://doi.org/10.3390/microplastics2010012