Multi-Objective Evolutionary Rule-Based Classification with Categorical Data
Abstract
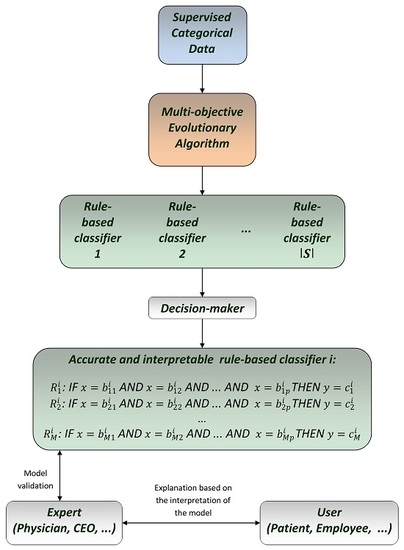
:1. Introduction
2. Background
2.1. Multi-Objective Constrained Optimization
2.2. The Multi-Objective Evolutionary Algorithms ENORA and NSGA-II
| Algorithm 1 strategy for multi-objective optimization. |
| Require: {Number of generations} Require: {Number of individuals in the population}
|
| Algorithm 2 Binary tournament selection. |
Require:P {Population}
|
| Algorithm 3 Rank-crowding function. |
| Require:P {Population} Require: {Individuals to compare}
|
| Algorithm 4 Crowding_distance function. |
| Require:P {Population} Require: P {Population} Require: I {Individual} Require: l {Number of objectives}
|
2.3. PART
2.4. JRip
2.5. OneR
2.6. ZeroR
3. Multi-Objective Optimization for Categorical Rule-Based Classification
3.1. Rule-Based Classification for Categorical Data
3.2. A Multi-Objective Optimization Solution
3.2.1. Representation
- Integer values associated to the antecedents , for and .
- Integer values associated to the consequent , for .
3.2.2. Constraint Handling
3.2.3. Initial Population
- Individuals are uniformly distributed with respect to the number of rules with values between w and , and with an additional constraint that specifies that there must be at least one individual for each number of rules (Steps 4–8). This ensures an adequate initial diversity in the search space in terms of the second objective of the optimization model.
- All individuals contain at least one rule for any output class between 1 and w (Steps 16–20).
| Algorithm 5 Initialize population. |
| Require: {Number of categorical input attributes} Require: , , {Number of categories for the input attributes} Require: , {Number of classes for the output attribute} Require: {Number of crossing operators} Require: {Number of mutation operators} Require: {Maximum number of rules} Require: {Number of individuals in the population}
|
3.2.4. Fitness Functions
3.2.5. Variation Operators
| Algorithm 6 Variation. |
Require:, {Individuals for variation}
|
| Algorithm 7 Self-adaptive crossover. |
| Require:I, J {Individuals for crossing} Require: () {Probability of variation} Require: {Number of different crossover operators}
|
| Algorithm 8 Self-adaptive mutation. |
| Require:I {Individual for mutation} Require: () {Probability of variation} Require: {Number of different mutation operators}
|
| Algorithm 9 Rule crossover. |
Require:I, J {Individuals for crossing}
|
| Algorithm 10 Rule incremental crossover. |
| Require:I, J {Individuals for crossing} Require: {Maximum number of rules}
|
| Algorithm 11 Rule incremental mutation. |
| Require:I {Individual for mutation} Require: {Maximum number of rules}
|
| Algorithm 12 Integer mutation. |
| Require:I {Individual for mutation} Require: {Number of categorical input attributes} Require: , , {Number of categories for the input attributes}
|
4. Experiment and Results
4.1. The Breast Cancer Dataset
4.2. The Monk’s Problem 2 Dataset
{heap_shape= round, body_shape=round, is_smiling=yes, holding=sword, jacket_color=red,
has_tie=yes}
4.3. Optimization Models
4.4. Choosing the Best Pareto Front
4.5. Comparing Our Method with Other Classifier Learning Systems (Full Training Mode)
4.6. Comparing Our Method with Other Classifier Learning Systems (Cross-Validation and Train/Test Percentage Split Mode)
4.7. Additional Experiments
- Tic-Tac-Toe-Endgame dataset, with 9 input attributes, 958 instances, and binary class (Table 11).
- Car dataset, with 6 input attributes, 1728 instances, and 4 output classes (Table 12).
- Chess (King-Rook vs. King-Pawn) (kr-vs-kp), with 36 input attributes, 3196 instances, and binary class (Table 13).
- Nursery dataset, with 8 input attributes, 12,960 instances, and 5 output classes (Table 14).
5. Analysis of Results and Discussion
6. Conclusions and Future Works
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ML | Machine learning |
| ANN | Artificial neural networks |
| DLNN | Deep learning neural networks |
| CEO | Chief executive officer |
| SVM | Support vector machines |
| IBL | Instance-based learning |
| DT | Decision trees |
| RBC | Rule-based classifiers |
| ROC | Receiver operating characteristic |
| RMSE | Root mean square error performance metric |
| FL | Fuzzy logic |
| MOEA | Multi-objective evolutionary algorithms |
| NSGA-II | Non-dominated sorting genetic algorithm, 2nd version |
| ENORA | Evolutionary non-dominated radial slots based algorithm |
| PART | Partial decision tree classifier |
| JRip | RIPPER classifier of Weka |
| RIPPER | Repeated incremental pruning to produce error reduction |
| OneR | One rule classifier |
| ZeroR | Zero rule classifier |
| ENORA-ACC | ENORA with objective function defined as accuracy |
| ENORA-AUC | ENORA with objective function defined as area under the ROC curve |
| ENORA-RMSE | ENORA with RMSE objective function |
| NSGA-II-ACC | NSGA-II with objective function defined as accuracy |
| NSGA-II-AUC | NSGA-II with objective function defined as area under the ROC curve |
| NSGA-II-RMSE | NSGA-II with RMSE objective function |
| HVR | Hypervolume ratio |
| TP | True positive |
| FP | False positive |
| MCC | Matthews correlation coefficient |
| PRC | Precision-recall curve |
Appendix A. Statistical Tests for Breast Cancer Dataset
| Algorithm | p-Value | Null Hypothesis |
|---|---|---|
| ENORA-ACC | 0.5316 | Not Rejected |
| ENORA-AUC | 0.3035 | Not Rejected |
| ENORA-RMSE | 0.7609 | Not Rejected |
| NSGA-II-ACC | 0.1734 | Not Rejected |
| NSGA-II-AUC | 0.3802 | Not Rejected |
| NSGA-II-RMSE | 0.6013 | Not Rejected |
| PART | 0.0711 | Not Rejected |
| JRip | 0.5477 | Not Rejected |
| OneR | 0.316 | Not Rejected |
| ZeroR | 3.818 × 10 | Rejected |
| p-Value | Null Hypothesis | |
|---|---|---|
| Friedman | 5.111 × 10 | Rejected |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip | OneR | |
|---|---|---|---|---|---|---|---|---|---|
| ENORA-AUC | 0.2597 | - | - | - | - | - | - | - | - |
| ENORA-RMSE | 0.9627 | 0.9627 | - | - | - | - | - | - | - |
| NSGA-II-ACC | 0.9981 | 0.8047 | 1.0000 | - | - | - | - | - | - |
| NSGA-II-AUC | 0.2951 | 1.0000 | 0.9735 | 0.8386 | - | - | - | - | - |
| NSGA-II-RMSE | 1.0000 | 0.2169 | 0.9436 | 0.9960 | 0.2486 | - | - | - | - |
| PART | 0.1790 | 1.0000 | 0.9186 | 0.6997 | 1.0000 | 0.1461 | - | - | - |
| JRip | 0.9909 | 0.8956 | 1.0000 | 1.0000 | 0.9186 | 0.9840 | 0.8164 | - | - |
| OneR | 0.0004 | 0.6414 | 0.0451 | 0.0108 | 0.5961 | 0.0002 | 0.7546 | 0.0212 | - |
| ZeroR | 0.2377 | 1.0000 | 0.9538 | 0.7803 | 1.0000 | 0.1973 | 1.0000 | 0.8783 | 0.6709 |
| ENORA-ACC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-RMSE | JRip | |
|---|---|---|---|---|---|
| OneR | ENORA-ACC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-RMSE | JRip |
| Algorithm | p-Value | Null Hypothesis |
|---|---|---|
| ENORA-ACC | 0.6807 | Not Rejected |
| ENORA-AUC | 0.3171 | Not Rejected |
| ENORA-RMSE | 0.6125 | Not Rejected |
| NSGA-II-ACC | 0.0871 | Not Rejected |
| NSGA-II-AUC | 0.5478 | Not Rejected |
| NSGA-II-RMSE | 0.6008 | Not Rejected |
| PART | 0.6066 | Not Rejected |
| JRip | 0.2978 | Not Rejected |
| OneR | 0.4531 | Not Rejected |
| ZeroR | 0.0000 | Rejected |
| p-Value | Null Hypothesis | |
|---|---|---|
| Friedman | 8.232 × 10 | Rejected |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip | OneR | |
|---|---|---|---|---|---|---|---|---|---|
| ENORA-AUC | 1.0000 | - | - | - | - | - | - | - | - |
| ENORA-RMSE | 0.9972 | 0.9990 | - | - | - | - | - | - | - |
| NSGA-II-ACC | 0.9999 | 1.0000 | 1.0000 | - | - | - | - | - | - |
| NSGA-II-AUC | 1.0000 | 1.0000 | 1.0000 | 1.0000 | - | - | - | - | - |
| NSGA-II-RMSE | 0.9990 | 0.9997 | 1.0000 | 1.0000 | 1.0000 | - | - | - | - |
| PART | 0.9999 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | - | - | - |
| JRip | 1.0000 | 1.0000 | 0.9992 | 1.0000 | 1.0000 | 0.9998 | 1.0000 | - | - |
| OneR | 0.0041 | 0.0062 | 0.0790 | 0.0323 | 0.0281 | 0.0582 | 0.0345 | 0.0067 | - |
| ZeroR | 3.8 × 10 | 7.2 × 10 | 4.6 × 10 | 9.8 × 10 | 7.8 × 10 | 2.7 × 10 | 1.1 × 10 | 8.1 × 10 | 0.6854 |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip | |
|---|---|---|---|---|---|---|---|---|
| OneR | ENORA-ACC | ENORA-AUC | - | NSGA-II-ACC | NSGA-II-AUC | - | PART | JRip |
| ZeroR | ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip |
| Algorithm | p-Value | Null Hypothesis |
|---|---|---|
| ENORA-ACC | 5.042 × 10 | Rejected |
| ENORA-AUC | 2.997 × 10 | Rejected |
| ENORA-RMSE | 4.762 × 10 | Rejected |
| NSGA-II-ACC | 4.88 × 10 | Rejected |
| NSGA-II-AUC | 2.339 × 10 | Rejected |
| NSGA-II-RMSE | 2.708 × 10 | Rejected |
| PART | 0.3585 | Not Rejected |
| JRip | 9.086 × 10 | Rejected |
| OneR | 1.007 × 10 | Rejected |
| ZeroR | 0.0000 | Rejected |
| p-Value | Null Hypothesis | |
|---|---|---|
| Friedman | 2.2 × 10 | Rejected |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip | OneR | |
|---|---|---|---|---|---|---|---|---|---|
| ENORA-AUC | 0.9998 | - | - | - | - | - | - | - | - |
| ENORA-RMSE | 0.0053 | 0.0004 | - | - | - | - | - | - | - |
| NSGA-II-ACC | 0.3871 | 0.0942 | 0.8872 | - | - | - | - | - | - |
| NSGA-II-AUC | 0.8872 | 0.4894 | 0.3871 | 0.9988 | - | - | - | - | - |
| NSGA-II-RMSE | 4.1 × 10 | 1.3 × 10 | 0.9860 | 0.2169 | 0.0244 | - | - | - | - |
| PART | 4.7 × 10 | 5.6 × 10 | 0.1973 | 0.0013 | 3.3 × 10 | 0.8689 | - | - | - |
| JRip | 0.2712 | 0.6997 | 1.2 × 10 | 7.0 × 10 | 0.0025 | 6.3 × 10 | 6.9 × 10 | - | - |
| OneR | 0.0062 | 0.0546 | 1.5 × 10 | 5.5 × 10 | 5.5 × 10 | 8.3 × 10 | 8.3 × 10 | 0.9584 | - |
| ZeroR | 1.9 × 10 | 0.0004 | 7.3 × 10 | 8.6 × 10 | 2.3 × 10 | 8.5 × 10 | <2 × 10 | 0.2377 | 0.9584 |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | |
|---|---|---|---|---|---|---|---|
| ENORA-RMSE | ENORA-ACC | NSGA-II-AUC | - | - | - | - | - |
| NSGA-II-RMSE | ENORA-ACC | ENORA-AUC | - | - | NSGA-II-AUC | - | - |
| PART | ENORA-ACC | ENORA-AUC | - | NSGA-II-ACC | NSGA-II-AUC | - | - |
| JRip | - | - | JRip | JRip | JRip | JRip | JRip |
| OneR | OneR | - | OneR | OneR | OneR | OneR | OneR |
| ZeroR | ZeroR | ZeroR | ZeroR | ZeroR | ZeroR | ZeroR | ZeroR |
Appendix B. Statistical Tests for Monk’s Problem 2 Dataset
| Algorithm | p-Value | Null Hypothesis |
|---|---|---|
| ENORA-ACC | 0.6543 | Not Rejected |
| ENORA-AUC | 0.6842 | Not Rejected |
| ENORA-RMSE | 0.0135 | Rejected |
| NSGA-II-ACC | 0.979 | Not Rejected |
| NSGA-II-AUC | 0.382 | Not Rejected |
| NSGA-II-RMSE | 0.0486 | Rejected |
| PART | 0.5671 | Not Rejected |
| JRip | 0.075 | Rejected |
| OneR | 4.672 × 10 | Rejected |
| ZeroR | 4.672 × 10 | Rejected |
| p-Value | Null Hypothesis | |
|---|---|---|
| Frideman | 1.292 × 10 | Rejected |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip | OneR | |
|---|---|---|---|---|---|---|---|---|---|
| ENORA-AUC | 0.8363 | - | - | - | - | - | - | - | - |
| ENORA-RMSE | 1.0000 | 0.9471 | - | - | - | - | - | - | - |
| NSGA-II-ACC | 0.1907 | 0.9902 | 0.3481 | - | - | - | - | - | - |
| NSGA-II-AUC | 0.0126 | 0.6294 | 0.0342 | 0.9958 | - | - | - | - | - |
| NSGA-II-RMSE | 0.0126 | 0.6294 | 0.0342 | 0.9958 | 1.0000 | - | - | - | - |
| PART | 0.8714 | 1.0000 | 0.9631 | 0.9841 | 0.5769 | 0.5769 | - | - | - |
| JRip | 2.1 × 10 | 0.0048 | 1.0 × 10 | 0.1341 | 0.6806 | 0.6806 | 0.0036 | - | - |
| OneR | 0.0001 | 0.0743 | 0.0006 | 0.6032 | 0.9875 | 0.9875 | 0.0601 | 0.9984 | - |
| ZeroR | 0.0001 | 0.0743 | 0.0006 | 0.6032 | 0.9875 | 0.9875 | 0.0601 | 0.9984 | 1.0000 |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | PART | |
|---|---|---|---|---|
| NSGA-II-AUC | ENORA-ACC | - | ENORA-RMSE | - |
| NSGA-II-RMSE | ENORA-ACC | - | ENORA-RMSE | - |
| JRip | ENORA-ACC | ENORA-AUC | ENORA-RMSE | PART |
| OneR | ENORA-ACC | - | ENORA-RMSE | - |
| ZeroR | ENORA-ACC | - | ENORA-RMSE | - |
| Algorithm | p-Value | Null Hypothesis |
|---|---|---|
| ENORA-ACC | 0.4318 | Not Rejected |
| ENORA-AUC | 0.7044 | Not Rejected |
| ENORA-RMSE | 0.0033 | Rejected |
| NSGA-II-ACC | 0.3082 | Not Rejected |
| NSGA-II-AUC | 0.0243 | Rejected |
| NSGA-II-RMSE | 0.7802 | Not Rejected |
| PART | 0.1641 | Not Rejected |
| JRip | 0.3581 | Not Rejected |
| OneR | 0.0000 | Rejected |
| ZeroR | 0.0000 | Rejected |
| p-Value | Null Hypothesis | |
|---|---|---|
| Frideman | 1.051 × 10 | Rejected |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip | OneR | |
|---|---|---|---|---|---|---|---|---|---|
| ENORA-AUC | 0.8363 | - | - | - | - | - | - | - | - |
| ENORA-RMSE | 1.0000 | 0.7054 | - | - | - | - | - | - | - |
| NSGA-II-ACC | 0.8870 | 0.0539 | 0.9556 | - | - | - | - | - | - |
| NSGA-II-AUC | 1.0000 | 0.8544 | 1.0000 | 0.8713 | - | - | - | - | - |
| NSGA-II-RMSE | 0.5504 | 0.0084 | 0.7054 | 0.9999 | 0.5239 | - | - | - | - |
| PART | 0.7054 | 1.0000 | 0.5504 | 0.0269 | 0.7295 | 0.0036 | - | - | - |
| JRip | 0.0238 | 2.3 × 10 | 0.0482 | 0.6806 | 0.0211 | 0.9471 | 7.0 × 10 | - | - |
| OneR | 0.0084 | 4.7 × 10 | 0.0186 | 0.4715 | 0.0073 | 0.8363 | 1.4 × 10 | 1.0000 | - |
| ZeroR | 0.0084 | 4.7 × 10 | 0.0186 | 0.4715 | 0.0073 | 0.8363 | 1.4 × 10 | 1.0000 | 1.0000 |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | PART | ||
|---|---|---|---|---|---|---|---|
| NSGA-II-RMSE | - | ENORA-AUC | - | - | - | - | - |
| PART | - | - | - | PART | - | PART | - |
| JRip | ENORA-ACC | ENORA-AUC | ENORA-RMSE | - | NSGA-II-AUC | - | PART |
| OneR | ENORA-ACC | ENORA-AUC | ENORA-RMSE | - | NSGA-II-AUC | - | PART |
| ZeroR | ENORA-ACC | ENORA-AUC | ENORA-RMSE | - | NSGA-II-AUC | - | PART |
| Algorithm | p-Value | Null Hypothesis |
|---|---|---|
| ENORA-ACC | 4.08 × 10 | Rejected |
| ENORA-AUC | 0.0002 | Rejected |
| ENORA-RMSE | 0.0094 | Rejected |
| NSGA-II-ACC | 0.0192 | Rejected |
| NSGA-II-AUC | 0.0846 | Rejected |
| NSGA-II-RMSE | 0.0037 | Rejected |
| PART | 0.9721 | Not Rejected |
| JRip | 0.0068 | Rejected |
| OneR | 0.0000 | Rejected |
| ZeroR | 0.0000 | Rejected |
| p-Value | Null Hypothesis | |
|---|---|---|
| Frideman | 2.657 × 10 | Rejected |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | JRip | OneR | |
|---|---|---|---|---|---|---|---|---|---|
| ENORA-AUC | 1.0000 | - | - | - | - | - | - | - | - |
| ENORA-RMSE | 1.0000 | 1.0000 | - | - | - | - | - | - | - |
| NSGA-II-ACC | 1.0000 | 1.0000 | 1.0000 | - | - | - | - | - | - |
| NSGA-II-AUC | 0.9925 | 0.9696 | 0.9984 | 0.9841 | - | - | - | - | - |
| NSGA-II-RMSE | 0.8870 | 0.9556 | 0.7966 | 0.9267 | 0.2622 | - | - | - | - |
| PART | 0.2824 | 0.1752 | 0.3957 | 0.2246 | 0.9015 | 0.0027 | - | - | - |
| JRip | 0.1752 | 0.2824 | 0.1110 | 0.2246 | 0.0084 | 0.9752 | 1.0 × 10 | - | - |
| OneR | 0.0211 | 0.0431 | 0.0110 | 0.0304 | 0.0004 | 0.6552 | 1.5 × 10 | 0.9993 | - |
| ZeroR | 0.0012 | 0.0031 | 0.0006 | 0.0020 | 1.0 × 10 | 0.1907 | 1.3 × 10 | 0.9015 | 0.9993 |
| ENORA-ACC | ENORA-AUC | ENORA-RMSE | NSGA-II-ACC | NSGA-II-AUC | NSGA-II-RMSE | PART | |
|---|---|---|---|---|---|---|---|
| PART | - | - | - | - | - | NSGA-II-RMSE | - |
| JRip | - | - | - | - | JRip | - | JRip |
| OneR | OneR | OneR | OneR | OneR | OneR | - | OneR |
| ZeroR | ZeroR | ZeroR | ZeroR | ZeroR | ZeroR | - | ZeroR |
Appendix C. Nomenclature
| Symbol | Definition |
|---|---|
| Equation (1): Multi-objective constrained optimization | |
| k-th decision variable | |
| Set of decision variables | |
| i-th objective function | |
| j-th constraint | |
| Number of objectives | |
| Number of constraints | |
| Number of decision variables | |
| Domain for each each decision variable | |
| Domain for the set of decision variables | |
| Set of all feasible solutions | |
| Set of non-dominated solutions or Pareto optimal set | |
| Pareto domination function | |
| Equation (2): Rule-based classification for categorical data | |
| Dataset | |
| categorical input attribute in the dataset | |
| Categorical input attributes in the dataset | |
| y | Categorical output attribute in the dataset |
| Domain of i-th categorical input attribute in the dataset | |
| Domain of categorical output attribute in the dataset | |
| Number of categorical input attributes in the dataset | |
| Rule-based classifier | |
| rule of classifier | |
| Category for categorical input attribute and rule of classifier | |
| Category for categorical output attribute and rule of classifier | |
| Compatibility degree of the rule of classifier for the example | |
| Result of the rule of classifier and categorical input attribute | |
| Association degree of classifier for the example with the class c | |
| Result of of the rule of classifier for the example with the class c | |
| Classification or output of the classifier for the example | |
| Equation (3): Multi-objective constrained optimization problem for rule-based classification | |
| Performance objective function of the classifier in the dataset | |
| Number of rules of the classifier | |
| Maximum number of rules allowed for classifiers | |
| Equations (4)–(6): Optimization models | |
| Acurracy: proportion of correctly classified instances with the classifier in the dataset | |
| K | Number of instances in the dataset |
| Result of the classification of the instance in the dataset with the classifier | |
| Predicted value of the instance in the dataset with the classifier | |
| Corresponding true value for the instance in the dataset . | |
| Area under the ROC curve obtained with the classifier in the dataset . | |
| Sensitivity: proportion of positive instances classified as positive with the classifier in the dataset | |
| Specificity: proportion of negative instances classified as negative with the classifier in the dataset | |
| t | Discrimination threshold |
| Square root of the mean square error obtained with the classifier in the dataset | |
| Equations (7) and (8): Hypervolume metric | |
| P | Population |
| Set of non-dominated individuals of P | |
| Volume of the search space dominated by the individual i | |
| Hypervolume: volume of the search space dominated by population P | |
| Volume of the search space non-dominated by population P | |
| Hypervolume ratio: ratio of over the volume of the entire search space | |
| Volume of the search space | |
| Minimum value for objective | |
| Maximum value for objective | |
| Minimum value for objective | |
| Maximum value for objective | |
References
- Bishop, C.M. Pattern Recognition and Machine Learning (Information Science and Statistics); Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach, 3rd ed.; Prentice Hall Press: Upper Saddle River, NJ, USA, 2009. [Google Scholar]
- Davalo, É. Neural Networks; MacMillan Computer Science; Macmillan Education: London, UK, 1991. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef] [Green Version]
- Aha, D.W.; Kibler, D.; Albert, M.K. Instance-based learning algorithms. Mach. Learn. 1991, 6, 37–66. [Google Scholar] [CrossRef] [Green Version]
- Gacto, M.; Alcalá, R.; Herrera, F. Interpretability of linguistic fuzzy rule-based systems: An overview of interpretability measures. Inf. Sci. 2011, 181, 4340–4360. [Google Scholar] [CrossRef]
- Cano, A.; Zafra, A.; Ventura, S. An EP algorithm for learning highly interpretable classifiers. In Proceedings of the 11th International Conference on Intelligent Systems Design and Applications, Cordoba, Spain, 22–24 November 2011; pp. 325–330. [Google Scholar]
- Liu, H.; Gegov, A. Collaborative Decision Making by Ensemble Rule Based Classification Systems. In Granular Computing and Decision-Making: Interactive and Iterative Approaches; Pedrycz, W., Chen, S.-M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 245–264. [Google Scholar]
- Sulzmann, J.N.; Fürnkranz, J. Rule Stacking: An Approach for Compressing an Ensemble of Rule Sets into a Single Classifier; Elomaa, T., Hollmén, J., Mannila, H., Eds.; Discovery Science; Springer: Berlin/Heidelberg, Germany, 2011; pp. 323–334. [Google Scholar]
- Jin, Y. Fuzzy Modeling of High-Dimensional Systems: Complexity Reduction and Interpretability Improvement. IEEE Trans. Fuzzy Syst. 2000, 8, 212–220. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Wadsworth and Brooks: Monterey, CA, USA, 1984. [Google Scholar]
- Novák, V.; Perfilieva, I.; Mockor, J. Mathematical Principles of Fuzzy Logic; Springer Science + Business Media: Heidelberg, Germany, 2012. [Google Scholar]
- Freund, Y.; Schapire, R.E. A Short Introduction to Boosting. In Proceedings of the Sixteenth International Joint Conference on Artificial Intelligence, Stockholm, Sweden, 31 July–6 August 1999; pp. 1401–1406. [Google Scholar]
- Deb, K. Multi-Objective Optimization Using Evolutionary Algorithms; John Wiley and Sons: London, UK, 2001. [Google Scholar]
- Coello, C.A.C.; van Veldhuizen, D.A.; Lamont, G.B. Evolutionary Algorithms for Solving Multi-Objective Problems; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2002. [Google Scholar]
- Jiménez, F.; Gómez-Skarmeta, A.; Sánchez, G.; Deb, K. An evolutionary algorithm for constrained multi-objective optimization. In Proceedings of the 2002 Congress on Evolutionary Computation, Honolulu, HI, USA, 12–17 May 2002; pp. 1133–1138. [Google Scholar]
- Jiménez, F.; Sánchez, G.; Juárez, J.M. Multi-objective evolutionary algorithms for fuzzy classification in survival prediction. Artif. Intell. Med. 2014, 60, 197–219. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, F.; Marzano, E.; Sánchez, G.; Sciavicco, G.; Vitacolonna, N. Attribute selection via multi-objective evolutionary computation applied to multi-skill contact center data classification. In Proceedings of the 2015 IEEE Symposium Series on Computational Intelligence, Cape Town, South Africa, 7–10 December 2015; pp. 488–495. [Google Scholar]
- Jiménez, F.; Jódar, R.; del Pilar Martín, M.; Sánchez, G.; Sciavicco, G. Unsupervised feature selection for interpretable classification in behavioral assessment of children. Expert Syst. 2017, 34, e12173. [Google Scholar] [CrossRef]
- Rey, M.; Galende, M.; Fuente, M.; Sainz-Palmero, G. Multi-objective based Fuzzy Rule Based Systems (FRBSs) for trade-off improvement in accuracy and interpretability: A rule relevance point of view. Knowl.-Based Syst. 2017, 127, 67–84. [Google Scholar] [CrossRef]
- Ducange, P.; Lazzerini, B.; Marcelloni, F. Multi-objective genetic fuzzy classifiers for imbalanced and cost-sensitive datasets. Soft Comput. 2010, 14, 713–728. [Google Scholar] [CrossRef]
- Gorzalczany, M.B.; Rudzinski, F. A multi-objective genetic optimization for fast, fuzzy rule-based credit classification with balanced accuracy and interpretability. Appl. Soft Comput. 2016, 40, 206–220. [Google Scholar] [CrossRef]
- Ducange, P.; Mannara, G.; Marcelloni, F.; Pecori, R.; Vecchio, M. A novel approach for internet traffic classification based on multi-objective evolutionary fuzzy classifiers. In Proceedings of the 2017 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), Naples, Italy, 9–12 July 2017; pp. 1–6. [Google Scholar]
- Antonelli, M.; Bernardo, D.; Hagras, H.; Marcelloni, F. Multiobjective Evolutionary Optimization of Type-2 Fuzzy Rule-Based Systems for Financial Data Classification. IEEE Trans. Fuzzy Syst. 2017, 25, 249–264. [Google Scholar] [CrossRef] [Green Version]
- Carmona, C.J.; González, P.; Deljesus, M.J.; Herrera, F. NMEEF-SD: Non-dominated multiobjective evolutionary algorithm for extracting fuzzy rules in subgroup discovery. IEEE Trans. Fuzzy Syst. 2010, 18, 958–970. [Google Scholar] [CrossRef]
- Hubertus, T.; Klaus, M.; Eberhard, T. Optimization Theory; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Sinha, S. Mathematical Programming: Theory and Methods; Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Collette, Y.; Siarry, P. Multiobjective Optimization: Principles and Case Studies; Springer-Verlag Berlin Heidelberg: New York, NY, USA, 2004. [Google Scholar]
- Karloff, H. Linear Programming; Birkhauser Basel: Boston, MA, USA, 1991. [Google Scholar]
- Maros, I.; Mitra, G. Simplex algorithms. In Advances in Linear and Integer Programming; Beasley, J.E., Ed.; Oxford University Press: Oxford, UK, 1996; pp. 1–46. [Google Scholar]
- Bertsekas, D. Nonlinear Programming, 2nd ed.; Athena Scientific: Cambridge, MA, USA, 1999. [Google Scholar]
- Jiménez, F.; Verdegay, J.L. Computational Intelligence in Theory and Practice. In Advances in Soft Computing; Reusch, B., Temme, K.-H., Eds.; Springer: Heidelberg, Germany, 2001; pp. 167–182. [Google Scholar]
- Jiménez, F.; Sánchez, G.; García, J.; Sciavicco, G.; Miralles, L. Multi-objective evolutionary feature selection for online sales forecasting. Neurocomputing 2017, 234, 75–92. [Google Scholar] [CrossRef]
- Deb, K.; Pratab, A.; Agarwal, S.; Meyarivan, T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 2002, 6, 182–197. [Google Scholar] [CrossRef] [Green Version]
- Bao, C.; Xu, L.; Goodman, E.D.; Cao, L. A novel non-dominated sorting algorithm for evolutionary multi-objective optimization. J. Comput. Sci. 2017, 23, 31–43. [Google Scholar] [CrossRef]
- Jiménez, F.; Sánchez, G.; Vasant, P. A Multi-objective Evolutionary Approach for Fuzzy Optimization in Production Planning. J. Intell. Fuzzy Syst. 2013, 25, 441–455. [Google Scholar]
- Deb, K.; Jain, H. An Evolutionary Many-Objective Optimization Algorithm Using Reference-Point-Based Nondominated Sorting Approach, Part I: Solving Problems With Box Constraints. IEEE Trans. Evol. Comput. 2014, 18, 577–601. [Google Scholar] [CrossRef]
- Frank, E.; Witten, I.H. Generating Accurate Rule Sets without Global Optimization; Department of Computer Science, University of Waikato: Waikato, New Zealand, 1998; pp. 144–151. [Google Scholar]
- Witten, I.H.; Frank, E.; Hall, M.A. Introduction to Weka. In Data Mining: Practical Machine Learning Tools and Techniques, 3rd ed.; Witten, I.H., Frank, E., Hall, M.A., Eds.; The Morgan Kaufmann Series in Data Management Systems; Morgan Kaufmann: Boston, MA, USA, 2011; pp. 403–406. [Google Scholar]
- Michalski, R.S. On the quasi-minimal solution of the general covering problem. In Proceedings of the V International Symposium on Information Processing (FCIP 69), Bled, Yugoslavia, 8–11 October 1969; pp. 125–128. [Google Scholar]
- Quinlan, J.R. C4. 5: Programs for Machine Learning; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Rajput, A.; Aharwal, R.P.; Dubey, M.; Saxena, S.; Raghuvanshi, M. J48 and JRIP rules for e-governance data. IJCSS 2011, 5, 201. [Google Scholar]
- Cohen, W.W. Fast effective rule induction. In Proceedings of the Twelfth International Conference on Machine Learning, Tahoe City, CA, USA, 9–12 July 1995; pp. 115–123. [Google Scholar]
- Fürnkranz, J.; Widmer, G. Incremental reduced error pruning. In Proceedings of the Eleventh International Conference, New Brunswick, NJ, USA, 10–13 July 1994; pp. 70–77. [Google Scholar]
- Holte, R.C. Very simple classification rules perform well on most commonly used datasets. Mach. Learn. 1993, 11, 63–90. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Maulik, U.; Bandyopadhyay, S.; Coello, C.A.C. A Survey of Multiobjective Evolutionary Algorithms for Data Mining: Part I. IEEE Trans. Evol. Comput. 2014, 18, 4–19. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Maulik, U.; Bandyopadhyay, S.; Coello, C.A.C. Survey of Multiobjective Evolutionary Algorithms for Data Mining: Part II. IEEE Trans. Evol. Comput. 2014, 18, 20–35. [Google Scholar] [CrossRef]
- Ishibuchi, H.; Murata, T.; Turksen, I. Single-objective and two-objective genetic algorithms for selecting linguistic rules for pattern classification problems. Fuzzy Sets Syst. 1997, 89, 135–150. [Google Scholar] [CrossRef]
- Srinivas, M.; Patnaik, L. Adaptive probabilities of crossover and mutation in genetic algorithms. IEEE Trans. Syst. Man Cybern B Cybern. 1994, 24, 656–667. [Google Scholar] [CrossRef]
- Zwitter, M.; Soklic, M. Breast Cancer Data Set. Yugoslavia. Available online: http://archive.ics.uci.edu/ml/datasets/Breast+Cancer (accessed on 5 September 2018).
- Thrun, S. MONK’s Problem 2 Data Set. Available online: https://www.openml.org/d/334 (accessed on 5 September 2018).
- Thrun, S.B.; Bala, J.; Bloedorn, E.; Bratko, I.; Cestnik, B.; Cheng, J.; Jong, K.D.; Dzeroski, S.; Fahlman, S.E.; Fisher, D.; et al. The MONK’s Problems A Performance Comparison of Different LearningAlgorithms. Available online: http://digilib.gmu.edu/jspui/bitstream/handle/1920/1685/91-46.pdf?sequence=1 (accessed on 5 September 2018).
- Metz, C.E. Basic principles of ROC analysis. Semin. Nucl. Med. 1978, 8, 283–298. [Google Scholar] [CrossRef]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Hand, D.J. Measuring classifier performance: A coherent alternative to the area under the ROC curve. Mach. Learn. 2009, 77, 103–123. [Google Scholar] [CrossRef]
- Zitzler, E.; Deb, K.; Thiele, L. Comparison of multiobjective evolutionary algorithms: Empirical results. Evol. Comput. 2000, 8, 173–195. [Google Scholar] [CrossRef] [PubMed]
- Zitzler, E.; Thiele, L.; Laumanns, M.; Fonseca, C.; Grunert da Fonseca, V. Performance Assessment of Multiobjective Optimizers: An Analysis and Review. IEEE Trans. Evol. Comput. 2002, 7, 117–132. [Google Scholar] [CrossRef]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence (II), Montreal, QC, Canada, 20–25 August 1995; pp. 1137–1143. [Google Scholar]
- Jiménez, F.; Jodár, R.; Sánchez, G.; Martín, M.; Sciavicco, G. Multi-Objective Evolutionary Computation Based Feature Selection Applied to Behaviour Assessment of Children. In Proceedings of the 9th International Conference on Educational Data Mining EDM 2016, Raleigh, NC, USA, 29 June–2 July 2016; pp. 1888–1897. [Google Scholar]




| Codification for Rule Set | Codification for Adaptive Crossing and Mutation | |||||
|---|---|---|---|---|---|---|
| Antecedents | Consequent | Associated Crossing | Associated Mutation | |||
| … | ||||||
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ||
| … | ||||||
| # | Attribute Name | Type | Possible Values |
|---|---|---|---|
| 1 | age | categorical | 10–19, 20–29, 30–39, 40–49, 50–59, 60–69, 70–79, 80–89, 90–99. |
| 2 | menopause | categorical | lt40, ge40, premeno |
| 3 | tumour-size | categorical | 0–4, 5–9, 10–14, 15–19, 20–24, 25–29, 30–34, 35–39, 40–44, 45–49, 50–54, 55–59 |
| 4 | inv-nodes | categorical | 0–2, 3–5, 6–8, 9–11, 12–14, 15–17, 18–20, 21–23, 24–26, 27–29, 30–32, 33–35, 36–39 |
| 5 | node-caps | categorical | yes, no |
| 6 | deg-malign | categorical | 1, 2, 3 |
| 7 | breast | categorical | left, right |
| 8 | breast-quad | categorical | left-up, left-low, right-up, right-low, central |
| 9 | irradiat | categorical | yes, no |
| 10 | class | categorical | no-recurrence-events, recurrence-events |
| # | Atttribute Name | Type | Possible Values |
|---|---|---|---|
| 1 | head_shape | categorical | round, square, octagon |
| 2 | body_shape | categorical | round, square, octagon |
| 3 | is_smiling | categorical | yes, no |
| 4 | holding | categorical | sword, balloon, flag |
| 5 | jacket_color | categorical | red, yellow, green, blue |
| 6 | has_tie | categorical | yes, no |
| 7 | class | categorical | yes, no |
| Method | Breast Cancer | Monk’s Problem 2 |
|---|---|---|
| ENORA-ACC | 244.92 s. | 428.14 s. |
| ENORA-AUC | 294.75 s. | 553.11 s. |
| ENORA-RMSE | 243.30 s. | 414.42 s. |
| NSGA-II-ACC | 127.13 s. | 260.83 s. |
| NSGA-II-AUC | 197.07 s. | 424.83 s. |
| NSGA-II-RMSE | 134.87 s. | 278.19 s. |
| Learning Model | Number of Rules | Percent Correct | TP Rate | FP Rate | Precision | Recall | F-Measure | MCC | ROC Area | PRC Area | RMSE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ENORA-ACC | 2 | 79.02 | 0.790 | 0.449 | 0.796 | 0.790 | 0.762 | 0.455 | 0.671 | 0.697 | 0.458 |
| ENORA-AUC | 2 | 75.87 | 0.759 | 0.374 | 0.751 | 0.759 | 0.754 | 0.402 | 0.693 | 0.696 | 0.491 |
| ENORA-RMSE | 2 | 77.62 | 0.776 | 0.475 | 0.778 | 0.776 | 0.744 | 0.410 | 0.651 | 0.680 | 0.473 |
| NSGA-II-ACC | 2 | 77.97 | 0.780 | 0.501 | 0.805 | 0.780 | 0.738 | 0.429 | 0.640 | 0.679 | 0.469 |
| NSGA-II-AUC | 2 | 75.52 | 0.755 | 0.368 | 0.749 | 0.755 | 0.752 | 0.399 | 0.693 | 0.696 | 0.495 |
| NSGA-II-RMSE | 2 | 79.37 | 0.794 | 0.447 | 0.803 | 0.794 | 0.765 | 0.467 | 0.673 | 0.700 | 0.454 |
| PART | 15 | 78.32 | 0.783 | 0.397 | 0.773 | 0.783 | 0.769 | 0.442 | 0.777 | 0.793 | 0.398 |
| JRip | 3 | 76.92 | 0.769 | 0.471 | 0.762 | 0.769 | 0.740 | 0.389 | 0.650 | 0.680 | 0.421 |
| OneR | 1 | 72.72 | 0.727 | 0.563 | 0.703 | 0.727 | 0.680 | 0.241 | 0.582 | 0.629 | 0.522 |
| ZeroR | - | 70.27 | 0.703 | 0.703 | 0.494 | 0.703 | 0.580 | 0.000 | 0.500 | 0.582 | 0.457 |
| Learning Model | Number of Rules | Percent Correct | TP Rate | FP Rate | Precision | Recall | F-Measure | MCC | ROC Area | PRC Area | RMSE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ENORA-ACC | 7 | 75.87 | 0.759 | 0.370 | 0.753 | 0.759 | 0.745 | 0.436 | 0.695 | 0.680 | 0.491 |
| ENORA-AUC | 7 | 68.71 | 0.687 | 0.163 | 0.836 | 0.687 | 0.687 | 0.523 | 0.762 | 0.729 | 0.559 |
| ENORA-RMSE | 7 | 77.70 | 0.777 | 0.360 | 0.777 | 0.777 | 0.762 | 0.481 | 0.708 | 0.695 | 0.472 |
| NSGA-II-ACC | 7 | 68.38 | 0.684 | 0.588 | 0.704 | 0.684 | 0.597 | 0.203 | 0.548 | 0.580 | 0.562 |
| NSGA-II-AUC | 7 | 66.38 | 0.664 | 0.175 | 0.830 | 0.664 | 0.661 | 0.497 | 0.744 | 0.715 | 0.580 |
| NSGA-II-RMSE | 7 | 68.71 | 0.687 | 0.591 | 0.737 | 0.687 | 0.595 | 0.226 | 0.548 | 0.583 | 0.559 |
| PART | 47 | 94.01 | 0.940 | 0.087 | 0.940 | 0.940 | 0.940 | 0.866 | 0.980 | 0.979 | 0.218 |
| JRip | 1 | 65.72 | 0.657 | 0.657 | 0.432 | 0.657 | 0.521 | 0.000 | 0.500 | 0.549 | 0.475 |
| OneR | 1 | 65.72 | 0.657 | 0.657 | 0.432 | 0.657 | 0.521 | 0.000 | 0.500 | 0.549 | 0.585 |
| ZeroR | - | 65.72 | 0.657 | 0.657 | 0.432 | 0.657 | 0.521 | 0.000 | 0.500 | 0.549 | 0.475 |
| Rule | Antecedents | Consequent | ||||||
|---|---|---|---|---|---|---|---|---|
| : | IF | age = 50–59 | AND | inv-nodes = 0–2 | AND | node-caps = no | ||
| AND | deg-malig = 1 | AND | breast = right | AND | breast-quad = left-low | THEN | class = no-recurrence-events | |
| : | IF | age = 60–69 | AND | inv-nodes = 18–20 | AND | node-caps = yes | ||
| AND | deg-malig = 3 | AND | breast = left | AND | breast-quad = right-up | THEN | class = recurrence-events | |
| Rule | Antecedents | Consequent | ||||||
|---|---|---|---|---|---|---|---|---|
| : | IF | head_shape = round | AND | body_shape = round | AND | is_smiling = no | ||
| AND | holding = sword | AND | jacket_color = red | AND | has_tie = yes | THEN | class = yes | |
| : | IF | head_shape = octagon | AND | body_shape = round | AND | is_smiling = no | ||
| AND | holding = sword | AND | jacket_color = red | AND | has_tie = no | THEN | class = yes | |
| : | IF | head_shape = round | AND | body_shape = round | AND | is_smiling = no | ||
| AND | holding = sword | AND | jacket_color = yellow | AND | has_tie = yes | THEN | class = yes | |
| : | IF | head_shape = round | AND | body_shape = round | AND | is_smiling = no | ||
| AND | holding = sword | AND | jacket_color = red | AND | has_tie = no | THEN | class = yes | |
| : | IF | head_shape = square | AND | body_shape = square | AND | is_smiling = yes | ||
| AND | holding = flag | AND | jacket_color = yellow | AND | has_tie = no | THEN | class = no | |
| : | IF | head_shape = octagon | AND | body_shape = round | AND | is_smiling = yes | ||
| AND | holding = balloon | AND | jacket_color = blue | AND | has_tie = no | THEN | class = no | |
| : | IF | head_shape = octagon | AND | body_shape = octagon | AND | is_smiling = yes | ||
| AND | holding = sword | AND | jacket_color = green | AND | has_tie = no | THEN | class = no | |
| Learning Model | Percent Correct | ROC Area | Serialized Model Size |
|---|---|---|---|
| ENORA-ACC | 73.45 | 0.61 | 9554.80 |
| ENORA-AUC | 70.16 | 0.62 | 9554.63 |
| ENORA-RMSE | 72.39 | 0.60 | 9557.77 |
| NSGA-II-ACC | 72.50 | 0.60 | 9556.20 |
| NSGA-II-AUC | 70.03 | 0.61 | 9555.70 |
| NSGA-II-RMSE | 73.34 | 0.60 | 9558.60 |
| PART | 68.92 | 0.61 | 55,298.13 |
| JRip | 71.82 | 0.61 | 7664.07 |
| OneR | 67.15 | 0.55 | 1524.00 |
| ZeroR | 70.30 | 0.50 | 915.00 |
| Learning Model | Percent Correct | ROC Area | Serialized Model Size |
|---|---|---|---|
| ENORA-ACC | 76.69 | 0.70 | 9586.50 |
| ENORA-AUC | 72.82 | 0.79 | 9589.30 |
| ENORA-RMSE | 75.66 | 0.68 | 9585.30 |
| NSGA-II-ACC | 70.07 | 0.59 | 9590.60 |
| NSGA-II-AUC | 67.08 | 0.70 | 9619.70 |
| NSGA-II-RMSE | 67.63 | 0.54 | 9565.90 |
| PART | 73.51 | 0.79 | 73,115.90 |
| JRip | 64.05 | 0.50 | 5956.90 |
| OneR | 65.72 | 0.50 | 1313.00 |
| ZeroR | 65.72 | 0.50 | 888.00 |
| # | Attribute Name | Type | Possible Values |
|---|---|---|---|
| 1 | top-left-square | categorical | x, o, b |
| 2 | top-middle-square | categorical | x, o, b |
| 3 | top-right-square | categorical | x, o, b |
| 4 | middle-left-square | categorical | x, o, b |
| 5 | middle-middle-square | categorical | x, o, b |
| 6 | middle-right-square | categorical | x, o, b |
| 7 | bottom-left-square | categorical | x, o, b |
| 8 | bottom-middle-square | categorical | x, o, b |
| 9 | bottom-right-square | categorical | x, o, b |
| 10 | class | categorical | positive, negative |
| # | Attribute Name | Type | Possible Values |
|---|---|---|---|
| 1 | buying | categorical | vhigh, high, med, low |
| 2 | maint | categorical | vhigh, high, med, low |
| 3 | doors | categorical | 2, 3, 4, 5-more |
| 4 | persons | categorical | 2, 4, more |
| 5 | lug_boot | categorical | small, med, big |
| 6 | safety | categorical | low, med, high |
| 7 | class | categorical | unacc, acc, good, vgood |
| # | Attribute Name | Type | Possible Values |
|---|---|---|---|
| 1 | bkblk | categorical | t, f |
| 2 | bknwy | categorical | t, f |
| 3 | bkon8 | categorical | t, f |
| 4 | bkona | categorical | t, f |
| 5 | bkspr | categorical | t, f |
| 6 | bkxbq | categorical | t, f |
| 7 | bkxcr | categorical | t, f |
| 8 | bkxwp | categorical | t, f |
| 9 | blxwp | categorical | t, f |
| 10 | bxqsq | categorical | t, f |
| 11 | cntxt | categorical | t, f |
| 12 | dsopp | categorical | t, f |
| 13 | dwipd | categorical | g, l |
| 14 | hdchk | categorical | t, f |
| 15 | katri | categorical | b, n, w |
| 16 | mulch | categorical | t, f |
| 17 | qxmsq | categorical | t, f |
| 18 | r2ar8 | categorical | t, f |
| 19 | reskd | categorical | t, f |
| 20 | reskr | categorical | t, f |
| 21 | rimmx | categorical | t, f |
| 22 | rkxwp | categorical | t, f |
| 23 | rxmsq | categorical | t, f |
| 24 | simpl | categorical | t, f |
| 25 | skach | categorical | t, f |
| 26 | skewr | categorical | t, f |
| 27 | skrxp | categorical | t, f |
| 28 | spcop | categorical | t, f |
| 29 | stlmt | categorical | t, f |
| 30 | thrsk | categorical | t, f |
| 31 | wkcti | categorical | t, f |
| 32 | wkna8 | categorical | t, f |
| 33 | wknck | categorical | t, f |
| 34 | wkovl | categorical | t, f |
| 35 | wkpos | categorical | t, f |
| 36 | wtoeg | categorical | n, t, f |
| 37 | class | categorical | won, nowin |
| # | Attribute Name | Type | Possible Values |
|---|---|---|---|
| 1 | parents | categorical | usual, pretentious, great_pret |
| 2 | has_nurs | categorical | proper, less_proper, improper, critical, very_crit |
| 3 | form | categorical | complete, completed, incomplete, foster |
| 4 | children | categorical | 1, 2, 3, more |
| 5 | housing | categorical | convenient, less_conv, critical |
| 6 | finance | categorical | convenient, inconv |
| 7 | social | categorical | nonprob, slightly_prob, problematic |
| 8 | health | categorical | recommended, priority, not_recom |
| 9 | class | categorical | not_recom, recommend, very_recom, priority, spec_prior |
| Learning Model | Number of Rules | Percent Correct | TP Rate | FP Rate | Precision | Recall | F-Measure | MCC | ROC Area | PRC Area | RMSE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Monk’s problem 2 | |||||||||||
| ENORA-ACC | 7 | 77.70 | 0.777 | 0.360 | 0.777 | 0.777 | 0.762 | 0.481 | 0.708 | 0.695 | 0.472 |
| PART | 47 | 79.53 | 0.795 | 0.253 | 0.795 | 0.795 | 0.795 | 0.544 | 0.884 | 0.893 | 0.380 |
| JRip | 1 | 62.90 | 0.629 | 0.646 | 0.526 | 0.629 | 0.535 | −0.034 | 0.478 | 0.537 | 0.482 |
| OneR | 1 | 65.72 | 0.657 | 0.657 | 0.432 | 0.657 | 0.521 | 0.000 | 0.500 | 0.549 | 0.586 |
| ZeroR | - | 65.72 | 0.657 | 0.657 | 0.432 | 0.657 | 0.521 | 0.000 | 0.491 | 0.545 | 0.457 |
| Tic-Tac-Toe-Endgame | |||||||||||
| ENORA-ACC/RMSE | 2 | 98.33 | 0.983 | 0.031 | 0.984 | 0.983 | 0.983 | 0.963 | 0.976 | 0.973 | 0.129 |
| PART | 49 | 94.26 | 0.943 | 0.076 | 0.942 | 0.943 | 0.942 | 0.873 | 0.974 | 0.969 | 0.220 |
| JRip | 9 | 97.81 | 0.978 | 0.031 | 0.978 | 0.978 | 0.978 | 0.951 | 0.977 | 0.977 | 0.138 |
| OneR | 1 | 69.94 | 0.699 | 0.357 | 0.701 | 0.699 | 0.700 | 0.340 | 0.671 | 0.651 | 0.548 |
| ZeroR | - | 65.35 | 0.653 | 0.653 | 0.427 | 0.653 | 0.516 | 0.000 | 0.496 | 0.545 | 0.476 |
| Car | |||||||||||
| ENORA-RMSE | 14 | 86.57 | 0.866 | 0.089 | 0.866 | 0.866 | 0.846 | 0.766 | 0.889 | 0.805 | 0.259 |
| PART | 68 | 95.78 | 0.958 | 0.016 | 0.959 | 0.958 | 0.958 | 0.929 | 0.990 | 0.979 | 0.1276 |
| JRip | 49 | 86.46 | 0.865 | 0.064 | 0.881 | 0.865 | 0.870 | 0.761 | 0.947 | 0.899 | 0.224 |
| OneR | 1 | 70.02 | 0.700 | 0.700 | 0.490 | 0.700 | 0.577 | 0.000 | 0.500 | 0.543 | 0.387 |
| ZeroR | - | 70.02 | 0.700 | 0.700 | 0.490 | 0.700 | 0.577 | 0.000 | 0.497 | 0.542 | 0.338 |
| kr-vs-kp | |||||||||||
| ENORA-RMSE | 10 | 94.87 | 0.949 | 0.050 | 0.950 | 0.949 | 0.949 | 0.898 | 0.950 | 0.927 | 0.227 |
| PART | 23 | 99.06 | 0.991 | 0.010 | 0.991 | 0.991 | 0.991 | 0.981 | 0.997 | 0.996 | 0.088 |
| JRip | 16 | 99.19 | 0.992 | 0.008 | 0.992 | 0.992 | 0.992 | 0.984 | 0.995 | 0.993 | 0.088 |
| OneR | 1 | 66.46 | 0.665 | 0.350 | 0.675 | 0.665 | 0.655 | 0.334 | 0.657 | 0.607 | 0.579 |
| ZeroR | - | 52.22 | 0.522 | 0.522 | 0.273 | 0.522 | 0.358 | 0.000 | 0.499 | 0.500 | 0.500 |
| Nursery | |||||||||||
| ENORA-ACC | 15 | 88.41 | 0.884 | 0.055 | 0.870 | 0.884 | 0.873 | 0.824 | 0.915 | 0.818 | 0.2153 |
| PART | 220 | 99.21 | 0.992 | 0.003 | 0.992 | 0.992 | 0.992 | 0.989 | 0.999 | 0.997 | 0.053 |
| JRip | 131 | 96.84 | 0.968 | 0.012 | 0.968 | 0.968 | 0.968 | 0.957 | 0.993 | 0.974 | 0.103 |
| OneR | 1 | 70.97 | 0.710 | 0.137 | 0.695 | 0.710 | 0.702 | 0.570 | 0.786 | 0.632 | 0.341 |
| ZeroR | - | 33.33 | 0.333 | 0.333 | 0.111 | 0.333 | 0.167 | 0.000 | 0.500 | 0.317 | 0.370 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez, F.; Martínez, C.; Miralles-Pechuán, L.; Sánchez, G.; Sciavicco, G. Multi-Objective Evolutionary Rule-Based Classification with Categorical Data. Entropy 2018, 20, 684. https://doi.org/10.3390/e20090684
Jiménez F, Martínez C, Miralles-Pechuán L, Sánchez G, Sciavicco G. Multi-Objective Evolutionary Rule-Based Classification with Categorical Data. Entropy. 2018; 20(9):684. https://doi.org/10.3390/e20090684
Chicago/Turabian StyleJiménez, Fernando, Carlos Martínez, Luis Miralles-Pechuán, Gracia Sánchez, and Guido Sciavicco. 2018. "Multi-Objective Evolutionary Rule-Based Classification with Categorical Data" Entropy 20, no. 9: 684. https://doi.org/10.3390/e20090684
APA StyleJiménez, F., Martínez, C., Miralles-Pechuán, L., Sánchez, G., & Sciavicco, G. (2018). Multi-Objective Evolutionary Rule-Based Classification with Categorical Data. Entropy, 20(9), 684. https://doi.org/10.3390/e20090684