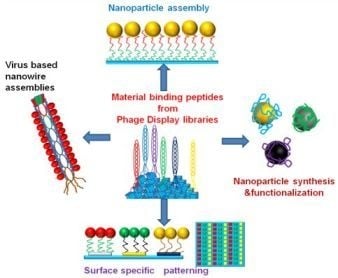
Material Binding Peptides for Nanotechnology
Abstract
:1. Introduction


2. Phage Display Selection and Screening of Material Binding Peptides

3. Selection and Characterization and Potential Uses of the Material Binding Peptides
3.1. Metal, Metal Oxide, Metal Alloys and Metal Compounds Binding Peptides




3.2. Semiconductor Binding Peptides
3.3. Mineral Binding Peptides
3.4. Carbon Materials Binding Peptides

3.5. Polymer Binding Peptides
4. Examples of Material Binding Peptides Utilization toward Practical Applications
| Material of Interest | Peptide Sequence | Notes |
|---|---|---|
| Gold | #VSGSSPDS [53], #LKAHLPPSRLPS [130] | Gold nanoparticle (NP)assembly |
| *TGTSVLIATPYV [56] | Gold NP synthesis | |
| Silver | *AYSSGAPPMPPF [131] | Ag NP synthesis |
| *IRPAIHIIPISH, *WSWRSPTPHVVT [58] | Ag NP synthesis | |
| Silica | #MSPHPHPRHHHT, #RGRRRRLSCRLL [74] | Silica precipitation |
| RLNPPSQMDPPF, QTWPPPLWFSTS [75] | SPR Keq(M−1): 0.12 × 106, 1.24 × 106 | |
| HPPMNASHPHMH, HTKHSHTSPPPL [132] | ||
| CHKKPSKSC [77] | LacI fusion QCM-D Keq (M−1): 2.46 × 108 [133] | |
| Titania/ Titanium | *RKLPDAPGMHTW [79,81] | Depletion assay Keq (M−1): 7.58 × 104 |
| *YPSAPPQWLTNT, *STPLVTGTNNLM *QSGSHVTGDLRL, *ATTLHPPRTSLP[87] | Subtractive biopanning | |
| #SCSDCLKSVDFIPSSLASS [73] | ELISA Keq(M−1): 4 × 106 | |
| #LNAAVPFTMAGS [92]. | ||
| #ATWVSPY [72] | Confocal microscopy | |
| *RKKRTKNPTHKLGGGW, *KSLSRHDHIHHHGGGW*TQHLSHPRYATKGGGW [84] | ||
| Zinc Oxide | *EAHVMHKVAPRP [89], *GLHVMHLVAPPR [90] | ZnO NP synthesis |
| *VRTRDDARTHRK [92] | Surface Quality Control | |
| Iridium Oxide | #AGETQQAM [93] | NP formation,co assembly |
| Iron Oxide | #LSTVQTISPSNH [95] | |
| Germania | *TGHQSPGAYAAH, *SLKMPHWPHLLP [94] | NP network formation |
| Platinum | *CPTSTGQAC, *CTLHVSSYC | SPR Keq(M−1): 3.4 × 106, 9.0 × 104, |
| Palladium | *QQSWPIS [134], *NFMSLPRLGHMH [69], | Pd NP synthesis |
| #SVTQNKY, #SPHPGPY, #HAPTPML [5] | Phage ELISA | |
| Aluminium | #VPSSGPQDTRTT, #YSPDPRPWSSRY [71] | |
| Stainless Steel | *MTWDPSLASPRS [92] | Surface Quality Control |
| *ATIHDAFYSAPE, *NLNPNTASAMHV [71] | ||
| Fe-Pt Alloy | #HNKHLPSTQPLA, SVSVGMKPSPRP, VISNHRESSRPL [96] | FePt NP synthesis |
| Cobalt | #HSVRWLLPGAHP, KLHSSPHTLPVQ, [58] | CoPt NP synthesis |
| Hydroxyapatite | #SVSVGMKPSPRP [109] | |
| *CMLPHHGAC [108] | Mineral synthesis | |
| Polymers | ||
| Poly(L-lactide) | *QLMHDYR [122] | SPR Keq (M−1): 6.1 × 104 |
| Polypyrrole | *THRTSTLDYFVI [119] | AFM analysis |
| it-PMMA | *ELWRPTR [135] | SPR Keq (M−1): 7.6 × 105 |
| sPS | #YLTMPTP | ELISA Keq(M−1): 2 × 1011 |
| Semiconductors | ||
| GaAs- InP | #AQNPSDNNTHTH [40], *SVSVGMKPSPRP [105] | |
| ZnS- PbS- CdS | #CNNPMHQNC, #QNPIHTH, #CTYSRLHLC [103] |
5. Conclusions
Acknowledgements
References
- Smith, G.P. Filamentous Fusion Phage - Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science 1985, 228, 1315–1317. [Google Scholar]
- Pande, J.; Szewczyk, M.M.; Grover, A.K. Phage display: Concept, innovations, applications and future. Biotechnol. Adv. 2010, 28, 849–858. [Google Scholar] [CrossRef]
- Rader, C.; Barbas, C.F. Phage display of combinatorial antibody libraries. Curr. Opin. Biotechnol. 1997, 8, 503–508. [Google Scholar] [CrossRef]
- Dunn, I.S. Phage display of proteins. Curr. Opin. Biotechnol. 1996, 7, 547–553. [Google Scholar] [CrossRef]
- Sarikaya, M.; Tamerler, C.; Jen, A.K.Y.; Schulten, K.; Baneyx, F. Molecular biomimetics: nanotechnology through biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar]
- Slocik, J.M.; Naik, R.R. Probing peptide-nanomaterial interactions. Chem. Soc. Rev. 2010, 39, 3454–3463. [Google Scholar] [CrossRef]
- Sethi, M.; Pacardo, D.B.; Knecht, M.R. Biological Surface Effects of Metallic Nanomaterials for Applications in Assembly and Catalysis. Langmuir 2010, 26, 15121–15134. [Google Scholar]
- Bratkovic, T. Progress in phage display: Evolution of the technique and its applications. Cell. Mol. Life Sci. 2010, 67, 749–767. [Google Scholar] [CrossRef]
- Huovinen, T.; Sanmark, H.; Yla-Pelto, J.; Vehniainen, M.; Lamminmaki, U. Oligovalent Fab Display on M13 Phage Improved by Directed Evolution. Mol. Biotechnol. 2010, 44, 221–231. [Google Scholar] [CrossRef]
- Alexander, P.A.; Rozak, D.A.; Orban, J.; Bryan, P.N. Directed evolution of highly homologous proteins with different folds by phage display: Implications for the protein folding code. Biochemistry 2005, 44, 14045–14054. [Google Scholar]
- Hu, Y. Biomimetic strategy for antifouling materials developed from mussel adhesive protein mimetic polymers. Mrs Bull. 2003, 28, 408–409. [Google Scholar] [CrossRef]
- Holzhuter, G.; Lakshminarayanan, K.; Gerber, T. Silica structure in the spicules of the sponge Suberites domuncula. Anal. Bioanal. Chem. 2005, 382, 1121–1126. [Google Scholar] [CrossRef]
- Yang, H.T.Y.; Lin, C.H.; Bridges, D.; Randall, C.J.; Hansma, P.K. Bio-inspired passive actuator simulating an abalone shell mechanism for structural control. Smart Mater. Struct. 2010, 19. [Google Scholar] [CrossRef]
- Mann, S. The biomimetics of enamel: A paradigm for organized biomaterials synthesis. Ciba Foundation Symp. 1997, 205, 261–274. [Google Scholar]
- Wang, X.H.; Schroder, H.C.; Muller, W.E.G. Giant Siliceous Spicules from the Deep-Sea Glass Sponge Monorhaphis Chuni. Int. Rev. Cell. Mol. Biol. 2009, 273, 69–115. [Google Scholar]
- Marin, F.; Luquet, G.; Marie, B.; Medakovic, D. Molluscan shell proteins: Primary structure, origin, and evolution. Curr. Top. Dev. Biol. 2008, 80, 209–276. [Google Scholar]
- Busch, S. Regeneration of human tooth enamel. Angew. Chem. Int. Ed. 2004, 43, 1428–1431. [Google Scholar] [CrossRef]
- Baldassarri, M.; Margolis, H.C.; Beniash, E. Compositional determinants of mechanical properties of enamel. J. Dent. Res. 2008, 87, 645–649. [Google Scholar] [CrossRef]
- Fong, H.; Foster, B.L.; Sarikaya, M.; Somerman, M.J. Structure and mechanical properties of Ank/Ank mutant mouse dental tissues--an animal model for studying periodontal regeneration. Arch. Oral Biol. 2009, 54, 570–576. [Google Scholar] [CrossRef]
- Metzler, R.A.; Evans, J.S.; Killian, C.E.; Zhou, D.; Churchill, T.H.; Appathurai, N.P.; Coppersmith, S.N.; Gilbert, P.U. Nacre protein fragment templates lamellar aragonite growth. J. Am. Chem. Soc. 2010, 132, 6329–6234. [Google Scholar]
- Lin, A.Y.; Chen, P.Y.; Meyers, M.A. The growth of nacre in the abalone shell. Acta Biomater. 2008, 4, 131–138. [Google Scholar] [CrossRef]
- Salih, E.; Wang, J.X.; Mah, J.; Fluckiger, R. Natural variation in the extent of phosphorylation of bone phosphoproteins as a function of in vivo new bone formation induced by demineralized bone matrix in soft tissue and bony environments. Biochem. J. 2002, 364, 465–474. [Google Scholar]
- Alves, N.M.; Leonor, I.B.; Azevedo, H.S.; Reis, R.L.; Mano, J.F. Designing biomaterials based on biomineralization of bone. J. Mater. Chem. 2010, 20, 2911–2921. [Google Scholar] [Green Version]
- Mahamid, J.; Aichmayer, B.; Shimoni, E.; Ziblat, R.; Li, C.H.; Siegel, S.; Paris, O.; Fratzl, P.; Weiner, S.; Addadi, L. Mapping amorphous calcium phosphate transformation into crystalline mineral from the cell to the bone in zebrafish fin rays. Proc. Natl. Acad. Sci. USA 2010, 107, 6316–6321. [Google Scholar]
- George, A.; Ravindran, S. Protein templates in hard tissue engineering. Nano Today 2010, 5, 254–266. [Google Scholar] [CrossRef]
- Lakshminarayanan, R.; Vivekanandan, S.; Samy, R.P.; Banerjee, Y.; Chi-Jin, E.O.; Teo, K.W.; Jois, S.D.S.; Kini, R.M.; Valiyaveettil, S. Structure, self-assembly, and dual role of a beta-defensin-like peptide from the chinese soft-shelled turtle eggshell matrix. J. Am. Chem. Soc. 2008, 130, 4660–4668. [Google Scholar]
- He, G.; Dahl, T.; Veis, A.; George, A. Nucleation of apatite crystals in vitro by self-assembled dentin matrix protein, 1. Nat. Mater. 2003, 2, 552–558. [Google Scholar] [CrossRef]
- Shen, X.Y.; Belcher, A.M.; Hansma, P.K.; Stucky, G.D.; Morse, D.E. Molecular cloning and characterization of lustrin A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J. Biol. Chem. 1997, 272, 32472–32481. [Google Scholar]
- Matsunaga, T.; Suzuki, T.; Tanaka, M.; Arakaki, A. Molecular analysis of magnetotactic bacteria and development of functional bacterial magnetic particles for nano-biotechnology. Trends Biotechnol. 2007, 25, 182–188. [Google Scholar]
- Gotliv, B.A.; Kessler, N.; Sumerel, J.L.; Morse, D.E.; Tuross, N.; Addadi, L.; Weiner, S. Asprich: A novel aspartic acid-rich protein family from the prismatic shell matrix of the bivalve Atrina rigida. Chembiochem 2005, 6, 304–314. [Google Scholar]
- Poulsen, N.; Sumper, M.; Kroger, N. Biosilica formation in diatoms: characterization of native silaffin-2 and its role in silica morphogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 12075–12080. [Google Scholar]
- Brown, S. Metal-recognition by repeating polypeptides. Nat. Biotechnol. 1997, 15, 269–272. [Google Scholar] [CrossRef]
- Brown, S. Engineered iron oxide-adhesion mutants of the Escherichia coli phage lambda receptor. Proc. Natl. Acad. Sci. USA 1992, 89, 8651–8655. [Google Scholar] [CrossRef]
- Sarikaya, M.; Tamerler, C.; Schwartz, D.T.; Baneyx, F.O. Materials assembly and formation using engineered polypeptides. Annu. Rev. Mater. Res. 2004, 34, 373–408. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar]
- Efimov, V.P.; Nepluev, I.V.; Mesyanzhinov, V.V. Bacteriophage-T4 as a Surface Display Vector. Virus Genes 1995, 10, 173–177. [Google Scholar] [CrossRef]
- Sternberg, N.; Hoess, R.H. Display of Peptides and Proteins on the Surface of Bacteriophage-Lambda. Proc. Natl. Acad. Sci. USA 1995, 92, 1609–1613. [Google Scholar] [CrossRef]
- Kriplani, U.; Kay, B.K. Selecting peptides for use in nanoscale materials using phagedisplayed combinatorial peptide libraries. Curr. Opin. Biotechnol. 2005, 16, 470–475. [Google Scholar] [CrossRef]
- Tamerler, C.; Sarikaya, M. Molecular biomimetics: nanotechnology and bionanotechnology using genetically engineered peptides. Phil. Trans. Roy. Soc. A-Math. Phys. Eng. Sci. 2009, 367, 1705–1726. [Google Scholar]
- Whaley, S.R.; English, D.S.; Hu, E.L.; Barbara, P.F.; Belcher, A.M. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature 2000, 405, 665–668. [Google Scholar]
- Sano, K.; Shiba, K. A hexapeptide motif that electrostatically binds to the surface of titanium. J. Am. Chem. Soc. 2003, 125, 14234–14235. [Google Scholar] [CrossRef]
- Park, T.J.; Lee, S.Y.; Lee, S.J.; Park, J.P.; Yang, K.S.; Lee, K.B.; Ko, S.; Park, J.B.; Kim, T.; Kim, S.K.; Shin, Y.B.; Chung, B.H.; Ku, S.J.; Kim, D.H.; Choi, I.S. Protein nanopatterns and biosensors using gold binding polypeptide as a fusion partner. Anal. Chem. 2006, 78, 7197–7205. [Google Scholar]
- Cesareni, G. Peptide Display on Filamentous Phage Capsids - a New Powerful Tool to Study Protein Ligand Interaction. FEBS Lett. 1992, 307, 66–70. [Google Scholar] [CrossRef]
- Hayashi, T.; Sano, K.; Shiba, K.; Kumashiro, Y.; Iwahori, K.; Yamashita, I.; Hara, M. Mechanism underlying specificity of proteins targeting inorganic materials. Nano Lett. 2006, 6, 515–519. [Google Scholar]
- Estephan, E.; Larroque, C.; Bec, N.; Martineau, P.; Cuisinier, F.J.; Cloitre, T.; Gergely, C. Selection and mass spectrometry characterization of peptides targeting semiconductor surfaces. Biotechnol. Bioeng. 2009, 104, 1121–1131. [Google Scholar] [CrossRef]
- Seker, U.O.; Wilson, B.; Sahin, D.; Tamerler, C.; Sarikaya, M. Quantitative affinity of genetically engineered repeating polypeptides to inorganic surfaces. Biomacromolecules 2009, 10, 250–257. [Google Scholar]
- Morita, Y.; Ohsugi, T.; Iwasa, Y.; Tamiya, E. A screening of phage displayed peptides for the recognition of fullerene (C60). J. Mol. Catal. B-Enzym. 2004, 28, 185–190. [Google Scholar] [CrossRef]
- Armitage, D.A.; Parker, T.L.; Grant, D.M. Biocompatibility and hemocompatibility of surface-modified NiTi alloys. J. Biomed. Mater. Res. Part A 2003, 66A, 129–137. [Google Scholar] [CrossRef]
- Colic, M.; Rudolf, R.; Stamenkovic, D.; Anzel, I.; Vucevic, D.; Jenko, M.; Lazic, V.; Lojen, G. Relationship between microstructure, cytotoxicity and corrosion properties of a Cu-Al-Ni shape memory alloy. Acta Biomater. 2010, 6, 308–317. [Google Scholar] [CrossRef]
- Nochomovitz, R.; Amit, M.; Matmor, M.; Ashkenasy, N. Bioassisted multi-nanoparticle patterning using single-layer peptide templates. Nanotechnology 2010, 21, 1–7. [Google Scholar]
- Brown, S.; Sarikaya, M.; Johnson, E. A genetic analysis of crystal growth. J. Mol. Biol. 2000, 299, 725–735. [Google Scholar] [CrossRef]
- Park, T.J.; Zheng, S.; Kang, Y.J.; Lee, S.Y. Development of a whole-cell biosensor by cell surface display of a gold-binding polypeptide on the gold surface. FEMS Microbiol. Lett. 2009, 293, 141–147. [Google Scholar] [CrossRef]
- Huang, Y.; Chiang, C.Y.; Lee, S.K.; Gao, Y.; Hu, E.L.; De Yoreo, J.; Belcher, A.M. Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Lett. 2005, 5, 1429–1434. [Google Scholar] [CrossRef]
- Slocik, J.M.; Stone, M.O.; Naik, R.R. Synthesis of gold nanoparticles using multifunctional peptides. Small 2005, 1, 1048–1052. [Google Scholar] [CrossRef]
- Naik, R.R.; Stringer, S.J.; Agarwal, G.; Jones, S.E.; Stone, M.O. Biomimetic synthesis and patterning of silver nanoparticles. Nat. Mater. 2002, 1, 169–172. [Google Scholar] [CrossRef]
- Kim, J.; Rheem, Y.; Yoo, B.; Chong, Y.; Bozhilov, K.N.; Kim, D.; Sadowsky, M.J.; Hur, H.G.; Myung, N.V. Peptide-mediated shape- and size-tunable synthesis of gold nanostructures. Acta Biomater. 2010, 6, 2681–2689. [Google Scholar]
- Lee, E.; Kim, D.H.; Woo, Y.; Hur, H.G.; Lim, Y. Solution structure of peptide AG4 used to form silver nanoparticles. Biochem. Biophys. Res. Commun. 2008, 376, 595–598. [Google Scholar]
- Naik, R.R.; Jones, S.E.; Murray, C.J.; McAuliffe, J.C.; Vaia, R.A.; Stone, M.O. Peptide templates for nanoparticle synthesis derived from polymerase chain reaction-driven phage display. Adv. Funct. Mater. 2004, 14, 25–30. [Google Scholar] [CrossRef]
- Sengupta, A.; Thai, C.K.; Sastry, M.S.R.; Matthaei, J.F.; Schwartz, D.T.; Davis, E.J.; Baneyx, F. A genetic approach for controlling the binding and orientation of proteins on nanoparticles. Langmuir 2008, 24, 2000–2008. [Google Scholar]
- Sarikaya, M.; Tamerler, C.; Jen, A.K.; Schulten, K.; Baneyx, F. Molecular biomimetics: nanotechnology through biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar]
- Kantarci, N.; Tamerler, C.; Sarikaya, M.; Haliloglu, T.; Doruker, P. Molecular dynamics simulations on constraint metal binding peptides. Polymer 2005, 46, 4307–4313. [Google Scholar] [CrossRef]
- Oren, E.E.; Tamerler, C.; Sarikaya, M. Metal recognition of septapeptides via polypod molecular architecture. Nano Lett. 2005, 5, 415–419. [Google Scholar] [CrossRef]
- Seker, U.O.S.; Wilson, B.; Dincer, S.; Kim, I.W.; Oren, E.E.; Evans, J.S.; Tamerler, C.; Sarikaya, M. Bn, Adsorption behavior of linear and cyclic genetically engineered platinum binding peptides. Langmuir 2007, 23, 7895–7900. [Google Scholar] [CrossRef]
- Seker, U.O.S.; Wilson, B.; Sahin, D.; Tamerler, C.; Sarikaya, M. Quantitative Affinity of Genetically Engineered Repeating Polypeptides to Inorganic Surfaces. Biomacromolecules 2009, 10, 250–257. [Google Scholar] [CrossRef]
- Dincer, S.; Tamerler, C.; Sarikaya, M.; Piskin, E. Photoresponsive peptide-azobenzene conjugates that specifically interact with platinum surfaces. Surf. Sci. 2008, 602, 1757–1762. [Google Scholar] [CrossRef]
- Khatayevich, D.; Gungormus, M.; Yazici, H.; So, C.; Cetinel, S.; Ma, H.; Jen, A.; Tamerler, C.; Sarikaya, M. Biofunctionalization of materials for implants using engineered peptides. Acta Biomater. 2010, 6, 4634–4641. [Google Scholar]
- Li, Y.; Whyburn, G.P.; Huang, Y. Specific peptide regulated synthesis of ultrasmall platinum nanocrystals. J. Am. Chem. Soc. 2009, 131, 15998–15999. [Google Scholar] [CrossRef]
- Sasaki, K.; Naohara, H.; Cai, Y.; Choi, Y.M.; Liu, P.; Vukmirovic, M.B.; Wang, J.X.; Adzic, R.R. Core-Protected Platinum Monolayer Shell High-Stability Electrocatalysts for Fuel-Cell Cathodes. Angew. Chem.-Int. Ed. 2010, 49, 8602–8607. [Google Scholar]
- Pacardo, D.B.; Sethi, M.; Jones, S.E.; Naik, R.R.; Knecht, M.R. Biomimetic synthesis of Pd nanocatalysts for the Stille coupling reaction. ACS Nano 2009, 3, 1288–1296. [Google Scholar] [CrossRef]
- Nian, R.; Kim, D.S.; Thuong, N.; Tan, L.H.; Kim, C.W.; Yoo, I.K.; Choe, W.S. Chromatographic biopanning for the selection of peptides with high specificity to Pb2+ from phage displayed peptide library. J. Chromatogr. A 2010, 1217, 5940–5949. [Google Scholar] [CrossRef]
- Zuo, R.J.; Ornek, D.; Wood, T.K. Aluminum- and mild steel-binding peptides from phage display. Appl. Microbiol. Biotechnol. 2005, 68, 505–509. [Google Scholar]
- Liu, Y.; Mao, J.; Zhou, B.; Wei, W.; Gong, S.Q. Peptide aptamers against titanium-based implants identified through phage display. J. Mater. Sci. Mater. M 2010, 21, 1103–1107. [Google Scholar]
- Meyers, S.R.; Hamilton, P.T.; Walsh, E.B.; Kenan, D.J.; Grinstaff, M.W. Endothelialization of titanium surfaces. Advan. Mater. 2007, 19, 2492–2498. [Google Scholar] [CrossRef]
- Naik, R.R.; Brott, L.L.; Clarson, S.J.; Stone, M.O. Silica-precipitating peptides isolated from a combinatorial phage display peptide library. J. Nanosci. Nanotechnol. 2002, 2, 95–100. [Google Scholar]
- Tamerler, C.; Kacar, T.; Sahin, D.; Fong, H.; Sarikaya, M. Genetically engineered polypeptides for inorganics: A utility in biological materials science and engineering. Mater. Sci. Eng. C-Biomim. Supramol. Syst. 2007, 27, 558–564. [Google Scholar] [CrossRef]
- Eteshola, E.; Brillson, L.J.; Lee, S.C. Selection and characteristics of peptides that bind thermally grown silicon dioxide films. Biomol. Eng. 2005, 22, 201–204. [Google Scholar] [CrossRef]
- Chen, H.B.; Su, X.D.; Neoh, K.G.; Choe, W.S. QCM-D analysis of binding mechanism of phage particles displaying a constrained heptapeptide with specific affinity to SiO2 and TiO2. Anal. Chem. 2006, 78, 4872–4879. [Google Scholar] [CrossRef]
- Chen, H.B.; Su, X.D.; Neoh, K.G.; Choe, W.S. Probing the interaction between peptides and metal oxides using point mutants of a TiO2-binding peptide. Langmuir 2008, 24, 6852–6857. [Google Scholar] [CrossRef]
- Sano, K.I.; Sasaki, H.; Shiba, K. Specificity and biomineralization activities of Ti-binding peptide-1 (TBP-1). Langmuir 2005, 21, 3090–3095. [Google Scholar] [CrossRef]
- Bonne, M.; Pronier, S.; Batonneau, Y.; Can, F.; Courtois, X.; Royer, S.; Marecot, P.; Duprez, D. Surface properties and thermal stability of SiO2-crystalline TiO2 nano-composites. J. Mater. Chem. 2010, 20, 9205–9214. [Google Scholar] [CrossRef]
- Sano, K.; Ajima, K.; Iwahori, K.; Yudasaka, M.; Iijima, S.; Yamashita, I.; Shiba, K. Endowing a ferritin-like cage protein with high affinity and selectivity for certain inorganic materials. Small 2005, 1, 826–832. [Google Scholar]
- Sano, K.; Sasaki, H.; Shiba, K. Utilization of the pleiotropy of a peptidic aptamer to fabricate heterogeneous nanodot-containing multilayer nanostructures. J. Am. Chem. Soc. 2006, 128, 1717–1722. [Google Scholar]
- Sano, K.; Yoshii, S.; Yamashita, I.; Shiba, K. In aqua structuralization of a three-dimensional configuration using biomolecules. Nano Lett. 2007, 7, 3200–3202. [Google Scholar] [CrossRef]
- Dickerson, M.B.; Jones, S.E.; Cai, Y.; Ahmad, G.; Naik, R.R.; Kroger, N.; Sandhage, K.H. Identification and design of peptides for the rapid, high-yield formation of nanoparticulate TiO2 from aqueous solutions at room temperature. Chem. Mater. 2008, 20, 1578–1584. [Google Scholar]
- Oren, E.E.; Tamerler, C.; Sahin, D.; Hnilova, M.; Seker, U.O.; Sarikaya, M.; Samudrala, R. A novel knowledge-based approach to design inorganic-binding peptides. Bioinformatics 2007, 23, 2816–2822. [Google Scholar] [CrossRef]
- Notman, R.; Oren, E.E.; Tamerler, C.; Sarikaya, M.; Samudrala, R.; Walsh, T.R. Solution Study of Engineered Quartz Binding Peptides Using Replica Exchange Molecular Dynamics. Biomacromolecules 2010, 11, 3266–3274. [Google Scholar] [CrossRef]
- Fang, Y.; Poulsen, N.; Dickerson, M.B.; Cai, Y.; Jones, S.E.; Naik, R.R.; Kroger, N.; Sandhage, K.H. Identification of peptides capable of inducing the formation of titania but not silica via a subtractive bacteriophage display approach. J. Mater. Chem. 2008, 18, 3871–3875. [Google Scholar]
- Ginley, D.S.; Bright, C. Transparent conducting oxides. Mrs Bull. 2000, 25, 15–18. [Google Scholar] [CrossRef]
- Umetsu, M.; Mizuta, M.; Tsumoto, K.; Ohara, S.; Takami, S.; Watanabe, H.; Kumagai, I.; Adschiri, T. Bioassisted room-temperature immobilization and mineralization of zinc oxide - The structural ordering of ZnO nanoparticles into a flower-type morphology. Advan. Mater. 2005, 17, 2571–2575. [Google Scholar] [CrossRef]
- Tomczak, M.M.; Gupta, M.K.; Drummy, L.F.; Rozenzhak, S.M.; Nalk, R.R. Morphological control and assembly of zinc oxide using a biotemplate. Acta Biomater. 2009, 5, 876–882. [Google Scholar] [CrossRef]
- He, R.L.; Tsuzuki, T. Low-Temperature Solvothermal Synthesis of ZnO Quantum Dots. J. Am. Ceram. Soc. 2010, 93, 2281–2285. [Google Scholar] [CrossRef]
- Vreuls, C.; Zocchi, G.; Genin, A.; Archambeau, C.; Martial, J.; Van de Weerdt, C. Inorganic-binding peptides as tools for surface quality control. J. Inorg. Biochem. 2010, 104, 1013–1021. [Google Scholar]
- Nam, Y.S.; Magyar, A.P.; Lee, D.; Kim, J.W.; Yun, D.S.; Park, H.; Pollom, T.S., Jr.; Weitz, D.A.; Belcher, A.M. Biologically templated photocatalytic nanostructures for sustained light-driven water oxidation. Nat. Nanotechnol. 2010, 5, 340–344. [Google Scholar]
- Dickerson, M.B.; Naik, R.R.; Stone, M.O.; Cai, Y.; Sandhage, K.H. Identification of peptides that promote the rapid precipitation of germania nanoparticle networks via use of a peptide display library. Chem. Commun. 2004, 1776–1777. [Google Scholar]
- Lower, B.H.; Lins, R.D.; Oestreicher, Z.; Straatsma, T.P.; Hochella, M.F.; Shi, L.A.; Lower, S.K. In vitro evolution of a peptide with a hematite binding motif that may constitute a natural metal-oxide binding archetype. Environ. Sci. Technol. 2008, 42, 3821–3827. [Google Scholar] [CrossRef]
- Reiss, B.D.; Mao, C.B.; Solis, D.J.; Ryan, K.S.; Thomson, T.; Belcher, A.M. Biological routes to metal alloy ferromagnetic nanostructures. Nano Lett. 2004, 4, 1127–1132. [Google Scholar] [CrossRef]
- Lee, S.K.; Yun, D.S.; Belcher, A.M. Cobalt ion mediated self-assembly of genetically engineered bacteriophage for biomimetic Co-Pt hybrid material. Biomacromolecules 2006, 7, 14–17. [Google Scholar] [CrossRef]
- Ahmad, G.; Dickerson, M.B.; Cai, Y.; Jones, S.E.; Ernst, E.M.; Vernon, J.P.; Haluska, M.S.; Fang, Y.; Wang, J.; Subrarnanyarn, G.; Naik, R.R.; Sandhage, K.H. Rapid bioenabled formation of ferroelectric BaTiO3 at room temperature from an aqueous salt solution at near neutral pH. J. Am. Chem. Soc. 2008, 130, 4–5. [Google Scholar]
- Reiss, B.D.; Bai, G.R.; Auciello, O.; Ocola, L.E.; Firestone, M.A. Identification of peptides for the surface functionalization of perovskite ferroelectrics. Appl. Phys. Lett. 2006, 88, 1–3. [Google Scholar]
- Nizamoglu, S.; Demir, H.V. Nanocrystal-based hybrid white light generation with tunable colour parameters. J. Opt. A-Pure Appl. Opt. 2007, 9, S419–S424. [Google Scholar] [CrossRef]
- Medintz, I.L.; Konnert, J.H.; Clapp, A.R.; Stanish, I.; Twigg, M.E.; Mattoussi, H.; Mauro, J.M.; Deschamps, J.R. A fluorescence resonance energy transfer-derived structure of a quantum dot-protein bioconjugate nanoassembly. Proc. Natl. Acad. Sci. USA 2004, 101, 9612–9617. [Google Scholar]
- Estephan, E.; Larroque, C.; Cuisinier, F.J.G.; Balint, Z.; Gergely, C. Tailoring GaN semiconductor surfaces with biomolecules. J. Phys. Chem. B 2008, 112, 8799–8805. [Google Scholar]
- Flynn, C.E.; Mao, C.B.; Hayhurst, A.; Williams, J.L.; Georgiou, G.; Iverson, B.; Belcher, A.M. Synthesis and organization of nanoscale II-VI semiconductor materials using evolved peptide specificity and viral capsid assembly. J. Mater. Chem. 2003, 13, 2414–2421. [Google Scholar] [CrossRef]
- Flynn, C.E.; Lee, S.W.; Peelle, B.R.; Belcher, A.M. Viruses as vehicles for growth, organization and assembly of materials. Acta Mater. 2003, 51, 5867–5880. [Google Scholar] [CrossRef]
- Estephan, E.; Saab, M.B.; Larroque, C.; Martin, M.; Olsson, F.; Lourdudoss, S.; Gergely, C. Peptides for functionalization of InP semiconductors. J. Colloid Interface Sci. 2009, 337, 358–363. [Google Scholar]
- Goede, K.; Busch, P.; Grundmann, M. Binding specificity of a peptide on semiconductor surfaces. Nano Lett. 2004, 4, 2115–2120. [Google Scholar]
- Fincham, A.G.; Simmer, J.P. Amelogenin proteins of developing dental enamel. Ciba Foundation Symp. 1997, 205, 118–134. [Google Scholar]
- Gungormus, M.; Fong, H.; Kim, I.W.; Evans, J.S.; Tamerler, C.; Sarikaya, M. Regulation of in vitro calcium phosphate mineralization by combinatorially selected hydroxyapatite-binding peptides. Biomacromolecules 2008, 9, 966–973. [Google Scholar] [CrossRef]
- Roy, M.D.; Stanley, S.K.; Amis, E.J.; Becker, M.L. Identification of a highly specific hydroxyapatite-binding peptide using phage display. Advan. Mater. 2008, 20, 1830–1836. [Google Scholar]
- Gaskin, D.J.H.; Starck, K.; Vulfson, E.N. Identification of inorganic crystal-specific sequences using phage display combinatorial library of short peptides: A feasibility study. Biotechnol. Lett. 2000, 22, 1211–1216. [Google Scholar]
- Li, C.M.; Botsaris, G.D.; Kaplan, D.L. Selective in vitro effect of peptides on calcium carbonate crystallization. Cryst. Growth Des. 2002, 2, 387–393. [Google Scholar] [CrossRef]
- Cao, Q.; Kim, H.S.; Pimparkar, N.; Kulkarni, J.P.; Wang, C.J.; Shim, M.; Roy, K.; Alam, M.A.; Rogers, J.A. Medium-scale carbon nanotube thin-film integrated circuits on flexible plastic substrates. Nature 2008, 454, 495–500. [Google Scholar] [CrossRef]
- Shannon, M.A.; Bohn, P.W.; Elimelech, M.; Georgiadis, J.G.; Marinas, B.J.; Mayes, A.M. Science and technology for water purification in the coming decades. Nature 2008, 452, 301–310. [Google Scholar]
- Chung, K.; Lee, C.H.; Yi, G.C. Transferable GaN Layers Grown on ZnO-Coated Graphene Layers for Optoelectronic Devices. Science 2010, 330, 655–657. [Google Scholar] [CrossRef]
- Miller, J.R.; Outlaw, R.A.; Holloway, B.C. Graphene Double-Layer Capacitor with ac Line-Filtering Performance. Science 2010, 329, 1637–1639. [Google Scholar]
- Wang, S.; Humphreys, E.S.; Chung, S.Y.; Delduco, D.F.; Lustig, S.R.; Wang, H.; Parker, K.N.; Rizzo, N.W.; Subramoney, S.; Chiang, Y.M.; Jagota, A. Peptides with selective affinity for carbon nanotubes. Nat. Mater. 2003, 2, 196–200. [Google Scholar] [CrossRef]
- Walsh, T.R.; Tomasio, S.M. Investigation of the influence of surface defects on peptide adsorption onto carbon nanotubes. Mol. Biosyst. 2010, 6, 1707–1718. [Google Scholar] [CrossRef]
- Kulp, J.L., 3rd; Shiba, K.; Evans, J.S. Probing the conformational features of a phage display polypeptide sequence directed against single-walled carbon nanohorn surfaces. Langmuir 2005, 21, 11907–11914. [Google Scholar] [CrossRef]
- Sanghvi, A.B.; Miller, K.P.; Belcher, A.M.; Schmidt, C.E. Biomaterials functionalization using a novel peptide that selectively binds to a conducting polymer. Nat. Mater. 2005, 4, 496–502. [Google Scholar]
- Serizawa, T.; Sawada, T.; Matsuno, H.; Matsubara, T.; Sato, T. A peptide motif recognizing a polymer stereoregularity. J. Am. Chem. Soc. 2005, 127, 13780–13781. [Google Scholar]
- Serizawa, T.; Sawada, T.; Matsuno, H. Highly specific affinities of short peptides against synthetic polymers. Langmuir 2007, 23, 11127–11133. [Google Scholar] [CrossRef]
- Matsuno, H.; Sekine, J.; Yajima, H.; Serizawa, T. Biological selection of peptides for poly(L-lactide) substrates. Langmuir 2008, 24, 6399–6403. [Google Scholar] [CrossRef]
- Serizawa, T.; Techawanitchai, P.; Matsuno, H. Isolation of peptides that can recognize syndiotactic polystyrene. Chembiochem 2007, 8, 989–993. [Google Scholar]
- Serizawa, T.; Sawada, T.; Kitayama, T. Peptide motifs that recognize differences in polymer-film surfaces. Angew. Chem.-Int. Ed. 2007, 46, 723–726. [Google Scholar]
- Serizawa, T.; Iida, K.; Matsuno, H.; Kurita, K. Cellulose-binding heptapeptides identified by phage display methods. Chem. Lett. 2007, 36, 988–989. [Google Scholar] [CrossRef]
- Nam, K.T.; Lee, Y.J.; Krauland, E.M.; Kottmann, S.T.; Belcher, A.M. Peptide-mediated reduction of silver ions on engineered biological scaffolds. ACS Nano 2008, 2, 1480–1486. [Google Scholar] [CrossRef]
- Togashi, T.; Yokoo, N.; Umetsu, M.; Ohara, S.; Naka, T.; Takami, S.; Abe, H.; Kumagai, I.; Adschiri, T. Material-binding peptide application-ZnO crystal structure control by means of a ZnO-binding peptide. J. Biosci. Bioeng. 2010, 111, 140–145. [Google Scholar]
- Donatan, S.; Yazici, H.; Bermek, H.; Sarikaya, M.; Tamerler, C.; Urgen, M. Physical elution in phage display selection of inorganic-binding peptides. Mater. Sci. Eng. C-Biomim. Supramol. Syst. 2009, 29, 14–19. [Google Scholar] [CrossRef]
- Hayashi, T.; Sano, K.I.; Shiba, K.; Iwahori, K.; Yamashita, I.; Hara, M. Critical Amino Acid Residues for the Specific Binding of the Ti-Recognizing Recombinant Ferritin with Oxide Surfaces of Titanium and Silicon. Langmuir 2009, 25, 10901–10906. [Google Scholar] [CrossRef]
- Nam, K.T.; Kim, D.W.; Yoo, P.J.; Chiang, C.Y.; Meethong, N.; Hammond, P.T.; Chiang, Y.M.; Belcher, A.M. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science 2006, 312, 885–888. [Google Scholar] [CrossRef]
- Naik, R.R.; Stringer, S.J.; Agarwal, G.; Jones, S.E.; Stone, M.O. Biomimetic synthesis and patterning of silver nanoparticles. Nat. Mater. 2002, 1, 169–172. [Google Scholar] [CrossRef]
- Eteshola, E.; Brillson, L.J.; Lee, S.C. Selection and characteristics of peptides that bind thermally grown silicon dioxide films. Biomol. Eng. 2005, 22, 201–204. [Google Scholar] [CrossRef]
- Chen, H.B.; Su, X.D.; Neoh, K.G.; Choe, W.S. Engineering Lacl for Self Assembly of Inorganic Nanoparticles on DNA Scaffold through the Understanding of Lacl Binding to Solid Surfaces. Adv. Funct. Mater. 2009, 19, 1186–1192. [Google Scholar]
- Chiu, C.Y.; Li, Y.J.; Huang, Y. Size-controlled synthesis of Pd nanocrystals using a specific multifunctional peptide. Nanoscale 2010, 2, 927–930. [Google Scholar] [CrossRef]
- Date, T.; Tanaka, K.; Nagamura, T.; Serizawa, T. Directional affinity of short peptides for synthetic polymers. Chem. Mater. 2008, 20, 4536–4538. [Google Scholar]
- Kacar, T.; Ray, J.; Gungormus, M.; Oren, E.E.; Tamerler, C.; Sarikaya, M. Quartz Binding Peptides as Molecular Linkers towards Fabricating Multifunctional Micropatterned Substrates. Advan. Mater. 2009, 21, 295–299. [Google Scholar]
- Zin, M.T.; Munro, A.M.; Gungormus, M.; Wong, N.Y.; Ma, H.; Tamerler, C.; Ginger, D.S.; Sarikaya, M.; Jen, A.K.Y. Peptide-mediated surface-immobilized quantum dot hybrid nanoassemblies with controlled photoluminescence. J. Mater. Chem. 2007, 17, 866–872. [Google Scholar]
- Cui, Y.; Kim, S.N.; Jones, S.E.; Wissler, L.L.; Naik, R.R.; McAlpine, M.C. Chemical Functionalization of Graphene Enabled by Phage Displayed Peptides. Nano Lett. 2010, 10, 4559–4565. [Google Scholar] [CrossRef]
- Kuang, Z.; Kim, S.N.; Crookes-Goodson, W.J.; Farmer, B.L.; Naik, R.R. Biomimetic chemosensor: designing peptide recognition elements for surface functionalization of carbon nanotube field effect transistors. ACS Nano 2010, 4, 452–458. [Google Scholar] [CrossRef]
- Mao, C.; Solis, D.J.; Reiss, B.D.; Kottmann, S.T.; Sweeney, R.Y.; Hayhurst, A.; Georgiou, G.; Iverson, B.; Belcher, A.M. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science 2004, 303, 213–217. [Google Scholar]
- Lee, Y.J.; Yi, H.; Kim, W.J.; Kang, K.; Yun, D.S.; Strano, M.S.; Ceder, G.; Belcher, A.M. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 2009, 324, 1051–1055. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Seker, U.O.S.; Demir, H.V. Material Binding Peptides for Nanotechnology. Molecules 2011, 16, 1426-1451. https://doi.org/10.3390/molecules16021426
Seker UOS, Demir HV. Material Binding Peptides for Nanotechnology. Molecules. 2011; 16(2):1426-1451. https://doi.org/10.3390/molecules16021426
Chicago/Turabian StyleSeker, Urartu Ozgur Safak, and Hilmi Volkan Demir. 2011. "Material Binding Peptides for Nanotechnology" Molecules 16, no. 2: 1426-1451. https://doi.org/10.3390/molecules16021426
APA StyleSeker, U. O. S., & Demir, H. V. (2011). Material Binding Peptides for Nanotechnology. Molecules, 16(2), 1426-1451. https://doi.org/10.3390/molecules16021426