Small-Molecule Inhibitors of the Type III Secretion System
Abstract
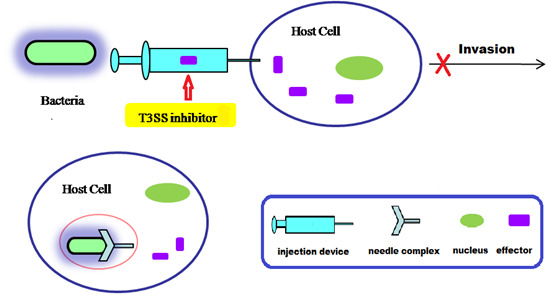
:1. Introduction
2. The Components of T3SS

3. Action Mechanism of T3SS

4. Small Molecule Inhibitors of T3SS
4.1. Salicylidene Acyl Hydrazides as T3SS Inhibitors

4.2. N-Hydroxybenzimidazoles as T3SS Inhibitors

4.3. Phenoxyacetamides as T3SS Inhibitors
| Compound | IC50 (μM) a | ||
|---|---|---|---|
| LcrF | ExsA b | SlyA c | |
 | 3.9 | 3.5 | >53.8 |
 | 8.0 | 5.9 | >55.2 |
 | 7.6 | 6.7 | >52 |



4.4. 2-Imino-5-arylidenethiazolidinones as T3SS Inhibitors

4.5. Plant Phenolic Compounds as T3SS Inhibitors

4.6. Polyol Products as T3SS Inhibitors

5. Conclusions and Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cegelski, L.; Marshall, G.R.; Eldridge, G.R.; Hultgren, S.J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008, 6, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Escaich, S. Antivirulene as a new antibacterial approach for chemotherapy. Curr. Opin. Chem. Biol. 2008, 12, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Sperandio, V. Anti-virulene strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Desvaux, M.; Hebraud, M.; Henderson, I.R.; Pallen, M.J. Type III secretion: What’s in a name? Trends Microbiol. 2006, 14, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Coburn, B.; Sekirov, I.; Finlay, B.B. Type III secretion systems and disease. Clin. Microbiol. Rev. 2007, 20, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.Y.; Xiao, Y.M.; Zhou, J.M. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant Microbe Interact. 2006, 19, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Waterman, S.R.; Holden, D.W. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 2003, 5, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.K.; Strynadka, N.C. New structural insights into the bacterial type III secretion system. Trends Biochem. Sci. 2006, 31, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Schraidt, O.; Marlovits, T.C. Three-dimensional model of Salmonella’s needle complex at subnamometer resolution. Science 2011, 331, 1192–1195. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Iida, T.; Park, K.S.; Goto, N.; Yasunaga, T.; Hiyoshi, H.; Matsuda, S.; Kodama, T.; Honda, T. Identification and characterization of a novel type III secretion system in trh-positive vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 2009, 77, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.R. The type III secretion injectisome, a complex nanomachine for intracellular “toxin” delivery. Biol. Chem. 2010, 391, 745–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.; Chaudhury, S.; McShan, A.C.; Kaur, K.; Guzman, N.D. Structure and biophysics of type III secretion in bacteria. Biochemistry 2013, 52, 2508–2517. [Google Scholar] [CrossRef] [PubMed]
- Tamano, K.; Aizawa, S.I.; Katayama, E.; Nonaka, T.; Imajoh-Ohmi, S.; Kuwae, A.; Nagai, S.; Sasakawa, C. Supramolecular structure of the Shigella type III secretion machinery: The needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000, 19, 3876–3887. [Google Scholar] [CrossRef] [PubMed]
- Spreter, T.; Yip, C.K.; Sanowar, S.; André, I.; Kimbrough, T.G.; Vuckovic, M.; Pfuetzner, R.A.; Deng, W.; Yu, A.C.; Finlay, B.B.; et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 2009, 16, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.E.; Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 2006, 444, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Loquet, A.; Sgourakis, N.G.; Gupta, R.; Giller, K.; Riedel, D.; Goosmann, C.; Griesinger, C.; Kolbe, M.; Baker, D.; Becker, S.; et al. Atomic model of the type III secretion system needle. Nature 2012, 486, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Akeda, Y.; Galan, J.E. Chaperone release and unfolding of substrates in type III secretion. Nature 2005, 437, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Izore, T.; Job, V.; Dessen, A. Biogenesis, regulation, and targeting of the type III secretion system. Structure 2011, 19, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.F.; Muller, S.; Wuest, E.; Cornelis, G.R. SycE allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol. Micrbiol. 2002, 46, 1183–1197. [Google Scholar] [CrossRef]
- Schraidt, O.; Lefebre, M.D.; Brunner, M.J.; Schmied, W.H.; Schmidt, A.; Radics, J.; Mechtler, K.; Galan, J.E.; Marlovits, T.C. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 2010, 6, e1000824. [Google Scholar] [CrossRef] [PubMed]
- Marlovits, T.C.; Kubori, T.; Sukhan, A.; Thomas, D.R.; Galan, J.E.; Unger, V.M. Structural insights into the assembly of the type III secretion needle complex. Science 2004, 306, 1040–1042. [Google Scholar] [CrossRef] [PubMed]
- Marlovits, T.C.; Kubori, T.; Lara-Tejero, M.; Thomas, D.; Unger, V.M.; Galan, J.E. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 2006, 441, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.E. Salmonella interactions with host cells: Type III secretion at work. Annu. Rev. Cell Dev. Biol. 2001, 17, 53–86. [Google Scholar] [CrossRef] [PubMed]
- Tsou, L.K.; Dossa, P.D.; Hang, H.C. Small molecules aimed at type III secretion systems to inhibit bacterial virulence. Med. Chem. Commun. 2013, 4, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Milla, C.E.; Chmiel, J.F.; Accurso, F.J.; van Devanter, D.R.; Konstan, M.W.; Yarranton, G.; Geller, D.E. Anti-PcrV antibody in cystic fibrosis: A novel approach targeting Pseudomonas aeruginosa airway infection. Pediatr. Pulmonol. 2014, 49, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, J.; Nordfelth, R.; Dubinina, E.; Bergman, T.; Gustafsson, M.; Magnusson, K.E.; Wolf-Watz, H. Modulation of virulence factor expression by pathogen target cell contact. Science 1996, 273, 1231–1233. [Google Scholar] [CrossRef] [PubMed]
- Kauppi, A.M.; Nordfelth, R.; Uvell, H.; Wolf-Watz, H.; Elofsson, M. Targeting bacterial virulence: Inhibitors of type III secretion in Yersinia. Chem. Biol. 2003, 10, 241–249. [Google Scholar] [CrossRef]
- Nordfelth, R.; Kauppi, A.M.; Norberg, H.A.; Wolf-Watz, H.; Elofsson, M. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 2005, 73, 3104–3114. [Google Scholar] [CrossRef] [PubMed]
- Keyser, P.; Elofsson, M.; Rosell, S.; Wolf-Watz, H. Virulence blockers as alternatives to antibiotics: Type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 2008, 264, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Muschiol, S.; Bailey, L.; Gylfe, A.; Sundin, C.; Hultenby, K.; Bergstro, S.; Elofsson, M.; Wolf-Watz, H.; Normark, S.; Normark, H.B. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 2006, 103, 14566–14571. [Google Scholar] [CrossRef]
- Gauthier, A.; Robertson, M.L.; Lowden, M.; Ibarra, J.A.; Puente, J.L.; Finlay, B.B. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 4101–4109. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.L.; Layton, A.N.; Field, T.R.; Bowen, A.J.; Wolf-Watz, H.; Elofsson, M.; Stevens, M.P.; Galyov, E.E. Inhibition of type III secretion in Salmonella enterica serovar typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 2007, 51, 2631–2635. [Google Scholar] [CrossRef] [PubMed]
- Hoe, N.P.; Minion, F.C.; Goguen, J.D. Temperature sensing in Yersinia pestis: Regulation of yopE transcription by lcrF. J. Bacteriol. 1992, 174, 4275–4286. [Google Scholar] [PubMed]
- Kim, O.K.; Garrity-Ryan, L.K.; Bartlett, V.J.; Grier, M.C.; Verma, A.K.; Medjanis, G.; Donatelli, J.E.; Macone, A.B.; Tanaka, S.K.; Levy, S.B.; et al. N-Hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: Novel antivirulence agents. J. Med. Chem. 2009, 52, 5626–5634. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.R.; Wolf-Watz, H. The Yersinia Yop virulon: A bacterial system for subverting eukaryotic cells. Mol. Microbiol. 1997, 23, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, L.K.; Mecsas, J. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infect. Immun. 2003, 71, 4595–4607. [Google Scholar] [CrossRef] [PubMed]
- Mota, L.J.; Cornelis, G.R. The bacterial injection kit: Type III secretion systems. Ann. Med. 2005, 37, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Monack, D.M.; Mecsas, J.; Bouley, D.; Falkow, S. Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J. Exp. Med. 1998, 188, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Hawley, R.J.; Eitzen, E.M. Biological weapons-a primer for microbiologists. Annu. Rev. Microbiol. 2001, 55, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, J.M.; Procter, J. Synthesis of N-alkoxybenzimidazoles with differentiated C2 and O-substituents. Tetrahedron Lett. 2001, 42, 5109–5111. [Google Scholar] [CrossRef]
- Ellison, D.W.; Miller, V.L. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 2006, 9, 153–159. [Google Scholar] [PubMed]
- Hong, M.; Fuangthong, M.; Helmann, J.D.; Brennan, R.G. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005, 20, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Garrity-Ryan, L.K.; Kim, O.K.; Balada-Llasat, J.M.; Bartlett, V.J.; Verma, A.K.; Fisher, M.L.; Castillo, C.; Songsungthong, W.; Tanaka, S.K.; Levy, S.B.; et al. Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model. Infect. Immun. 2010, 78, 4683–4690. [Google Scholar] [CrossRef] [PubMed]
- Shaver, C.M.; Hauser, A.R. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 2004, 72, 6969–6977. [Google Scholar] [CrossRef] [PubMed]
- Aiello, D.; Williams, J.D.; Majgier-Baranowska, H.; Patel, I.; Peet, N.P.; Huang, J.; Lory, S.; Bowlin, T.L.; Moir, D.T. Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 2010, 54, 1988–1999. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.F.; Wu, X.G.; Li, Yan; Liang, C.R.; Che, Y.Z.; Gu, L.L.; Ren, J.; Hu, K.; Sun, X.Q.; Yang, C.H.; et al. Synthesis and bioactivity of novel inhibitors for type III secretion system of Pseudomonas aeruginosa PAO1. Chin. J. Org. Chem. 2013, 33, 1309–1318. [Google Scholar] [CrossRef]
- Felise, H.B.; Nguyen, H.V.; Pfuetzner, R.A.; Barry, K.C.; Jackson, S.R.; Blanc, M.P.; Bronstein, P.A.; Kline, T.; Miller, S.I. An inhibitor of Gram-negative bacterial virulence protein secretion. Cell Host Microbe 2008, 4, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Kline, T.; Felise, H.B.; Barry, K.C.; Jackson, S.R.; Nguyen, H.V.; Miller, S.I. Substituted 2-imino-5-arylidenethiazolidin-4-one inhibitors of bacterial type III secretion. J. Med. Chem. 2008, 51, 7065–7074. [Google Scholar] [CrossRef] [PubMed]
- Kline, T.; Barry, K.C.; Jackson, S.R.; Felise, H.B.; Nguyen, H.V.; Miller, S.I. Tethered thiazolidinone dimers as inhibitors of the bacterial type III secretion system. Bioorg. Med. Chem. Lett. 2009, 19, 1340–1343. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Akashi, T.; Ayabe, S.I. Flavonoids of leguminous plants: Structure, biological activity, and biosynthesis. J. Plant Res. 2000, 113, 475–488. [Google Scholar] [CrossRef]
- Li, Y.; Peng, Q.; Selimi, D.; Wang, Q.; Charkowski, A.O.; Chen, X.; Yang, C.H. The plant phenolic compound p-coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl. Enviro. Microbiol. 2009, 75, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hutchins, W.C.; Wu, X.; Liang, C.; Zhang, C.; Yuan, X.; Khokhani, D.; Chen, X.; Che, Y.; Wang, Q.; et al. Derivatives of plant phenolic compound inhibits the type III secretion system of Dickeya dadantii via HrpX/HrpY two-component signal transduction and Rsm system. Mol. Plant Pathol. 2015, 16, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, A.; Li, J.; Zeng, Q.; Khokhani, D.; Hutchins, W.C.; Yost, A.C.; Biddle, E.; Toone, E.J.; Chen, X.; Yang, C.H. Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS/GacA two component signal transduction system. Antimicrob. Agents Chemother. 2012, 56, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Norelli, J.L.; Jones, A.L.; Aldwinckle, H.S. Fire blight management in the twenty-first century: Using new technologies that enhance host resistance in apple. Plant Dis. 2003, 87, 756–765. [Google Scholar] [CrossRef]
- McManus, P.S.; Stockwell, V.O.; Sundin, G.W.; Jones, A.L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 2002, 40, 443–463. [Google Scholar] [CrossRef] [PubMed]
- Khokhani, D.; Zhang, C.F.; Li, Y.; Wang, Q.; Zeng, Q.; Yamazaki, A.; Hutchins, W.; Zhou, S.S.; Chen, X.; Yang, C.H. Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 2013, 79, 5424–5436. [Google Scholar] [CrossRef] [PubMed]
- Grado, M.; Rosenberger, C.M.; Gauthier, A.; Vallance, B.A.; Finlay, B.B. Enteropathogenic Escherichia coli infection induces expression of the early growth response factor by activating mitogen-activated protein kinase cascades in epithelial cells. Infect. Immun. 2001, 69, 6217–6224. [Google Scholar] [CrossRef] [PubMed]
- Linington, R.G.; Robertson, M.; Gauthier, A.; Finlay, B.B.; van Soest, R.; Andersen, R.J. Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org. Lett. 2002, 4, 4089–4092. [Google Scholar] [CrossRef] [PubMed]
- Linington, R.G.; Robertson, M.; Gauthier, A.; Finlay, B.B.; MacMillan, J.B.; Molinski, T.F.; van Soest, R.; Andersen, R.J. Caminosides B-D, antimicrobial glycolipids isolated from the marine sponge Caminus sphaeroconia. J. Nat. Prod. 2006, 69, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Uchida, R.; Yoshijima, H.; Ui, H.; Shiomi, K.; Matsumoto, A.; Takahashi, Y.; Abe, A.; Tomoda, H.; Omura, S. Guadinomines, type III secretion system inhibitors, produced by Streptomyces sp. K01-0509 I. Taxonomy, fermentation, isolation and biological properties. J. Antibiot. 2008, 61, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Uchida, R.; Yoshijima, H.; Ui, H.; Shiomi, K.; Kim, Y.P.; Hirose, T.; Sunazuka, T.; Abe, A.; Tomoda, H.; et al. Guadinomines, type III secretion system inhibitors, produced by Streptomyces sp. K01-0509 II. Physico-chemical properties and structure elucidation. J. Antibiot. 2008, 61, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Iwatsuki, M.; Nagai, T.; Matsumoto, A.; Takahashi, Y.; Shiomi, K.; Omura, S.; Abe, A. A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. J. Antibiot. 2011, 64, 197–203. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, L.; Zhou, S.; Zhu, L.; Liang, C.; Chen, X. Small-Molecule Inhibitors of the Type III Secretion System. Molecules 2015, 20, 17659-17674. https://doi.org/10.3390/molecules200917659
Gu L, Zhou S, Zhu L, Liang C, Chen X. Small-Molecule Inhibitors of the Type III Secretion System. Molecules. 2015; 20(9):17659-17674. https://doi.org/10.3390/molecules200917659
Chicago/Turabian StyleGu, Lingling, Shanshan Zhou, Lanping Zhu, Cuirong Liang, and Xin Chen. 2015. "Small-Molecule Inhibitors of the Type III Secretion System" Molecules 20, no. 9: 17659-17674. https://doi.org/10.3390/molecules200917659
APA StyleGu, L., Zhou, S., Zhu, L., Liang, C., & Chen, X. (2015). Small-Molecule Inhibitors of the Type III Secretion System. Molecules, 20(9), 17659-17674. https://doi.org/10.3390/molecules200917659