Influences of Adhesion Variability on the “Living” Dynamics of Filamentous Bacteria in Microfluidic Channels
Abstract
:1. Introduction
2. Results
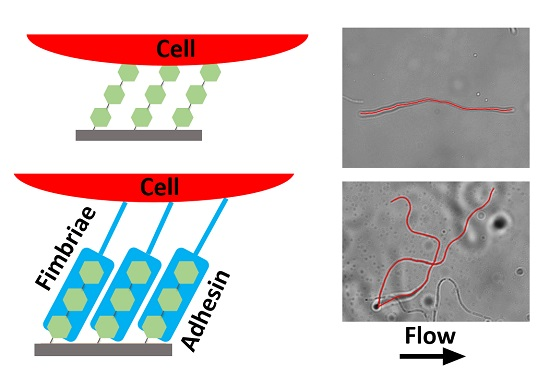
2.1. Initial Alignment of Adhered Filamentous Bacteria by Shear
2.2. Filament Elongation on Substrates with Different Affinity
2.3. Dynamic Geometry of Filamentous Bacteria during Elongation
3. Materials and Methods
3.1. Cell Culturing and Filamentation
3.2. Device Fabrication
3.3. Surface Modification
3.4. Experimental Setup for Flow Experiments
3.5. Image Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Martel, S. Bacterial microsystems and microrobots. Biomed. Microdevices 2012, 14, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.W. Biological Soft Robotics. Annu. Rev. Biomed. Eng. 2015, 17, 243–265. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Li, H.; Gu, T. A state of the art review on microbial fuel cells: A promising technology for wastewater treatment and bioenergy. Biotechnol. Adv. 2007, 25, 464–482. [Google Scholar] [CrossRef] [PubMed]
- Seker, U.O.; Demir, H.V. Material binding peptides for nanotechnology. Molecules 2011, 16, 1426–1451. [Google Scholar] [CrossRef] [PubMed]
- Justice, S.S.; Hunstad, D.A.; Seed, P.C.; Hultgren, S.J. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl. Acad. Sci. USA 2006, 103, 19884–19889. [Google Scholar] [CrossRef] [PubMed]
- Klein, K.; Palarasah, Y.; Kolmos, H.J.; Møller-Jensen, J.; Andersen, T.E. Quantification of filamentation by uropathogenic Escherichia coli during experimental bladder cell infection by using semi-automated image analysis. J. Microbiol. Methods 2015, 109, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Kougia, E.; Tselepi, M.; Vasilopoulos, G.; Lainioti, G.Ch.; Koromilas, N.D.; Druvari, D.; Bokias, G.; Vantarakis, A.; Kallitsis, J.K. Evaluation of Antimicrobial Efficiency of New Polymers Comprised by Covalently Attached and/or Electrostatically Bound Bacteriostatic Species, Based on Quaternary Ammonium Compounds. Molecules 2015, 20, 21313–21327. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Roux, A.; Ghigo, J.M. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 249–289. [Google Scholar] [PubMed]
- Hultgren, S.J.; Abraham, S.; Caparon, M.; Falk, P.; St Geme, J.W., 3rd; Normark, S. Pilus and nonpilus bacterial adhesins: Assembly and function in cell recognition. Cell 1993, 73, 887–901. [Google Scholar] [CrossRef]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Connell, I.; Agace, W.; Klemm, P.; Schembri, M.; Mărild, S.; Svanborg, C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 1996, 93, 9827–9832. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.E.; Trintchina, E.; Forero, M.; Vogel, V.; Sokurenko, E.V. Bacterial Adhesion to Target Cells Enhanced by Shear Force. Cell 2002, 109, 913–923. [Google Scholar] [CrossRef]
- Bouckaert, J.; Mackenzie, J.; de Paz, J.L.; Chipwaza, B.; Choudhury, D.; Zavialov, A.; Mannerstedt, K.; Anderson, J.; Piérard, D.; Wyns, L.; et al. The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of Escherichia coli pathotypes. Mol. Microbiol. 2006, 61, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Cabeen, M.T.; Jacobs-Wagner, C. Bacterial cell shape. Nat. Rev. Microbiol. 2005, 3, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.C.; Blair, K.M.; Salama, N.R. Staying in Shape: The Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Slater, M.; Schaechter, M. Control of cell division in bacteria. Bacteriol. Rev. 1974, 38, 199–221. [Google Scholar] [PubMed]
- Kawarai, T.; Wachi, M.; Ogino, H.; Furukawa, S.; Suzuki, K.; Ogihara, H.; Yamasaki, M. SulA-independent filamentation of Escherichia coli during growth after release from high hydrostatic pressure treatment. Appl. Microbiol. Biotechnol. 2003, 64, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Trusca, D.; Scott, S.; Thompson, C.; Bramhill, D. Bacterial SOS Checkpoint Protein SulA Inhibits Polymerization of Purified FtsZ Cell Division Protein. J. Bacteriol. 1998, 180, 3946–3953. [Google Scholar] [PubMed]
- Addinall, S.G.; Cao, C.; Lutkenhaus, J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 1997, 179, 4277–4284. [Google Scholar] [PubMed]
- Visvalingam, J.; Gill, C.O.; Holley, R.A. The viabilities of cells in cultures of Escherichia coli growing with formation of filaments at 6 degrees C. Int. J. Food Microbiol. 2012, 153, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.; Zhang, Q.; Vyawahare, S.; Rogers, E.; Rosenberg, S.M.; Austin, R.H. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc. Natl. Acad. Sci. USA 2015, 112, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Justice, S.S.; Hung, C.; Theriot, J.A.; Fletcher, D.A.; Anderson, G.G.; Footer, M.J.; Hultgren, S.J. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Schilling, J.D.; Mulvey, M.A.; Hultgren, S.J. Dynamic interactions between host and pathogen during acute urinary tract infections. Urology 2001, 57 (Suppl. 1), 56–61. [Google Scholar] [CrossRef]
- Möller, J.; Luehmann, T.; Hall, H.; Vogel, V. The race to the pole: How high-aspect ratio shape and heterogeneous environments limit phagocytosis of filamentous Escherichia coli bacteria by macrophages. Nano Lett. 2012, 12, 2901–2905. [Google Scholar] [CrossRef] [PubMed]
- Diver, J.M. Quinolone uptake by bacteria and bacterial killing. Rev. Infect. Dis. 1989, 11 (Suppl. 5), S941–S946. [Google Scholar] [CrossRef] [PubMed]
- Spratt, B.G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 1975, 72, 2999–3003. [Google Scholar] [CrossRef] [PubMed]
- Arends, S.J.; Weiss, D.S. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. J. Bacteriol. 2004, 186, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Jaimes-Lizcano, Y.A.; Hunn, D.D.; Papadopoulos, K.D. Filamentous Escherichia coli cells swimming in tapered microcapillaries. Res. Microbiol. 2014, 165, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Maki, N.; Gestwicki, J.E.; Lake, E.M.; Kiessling, L.L.; Adler, J. Motility and Chemotaxis of Filamentous Cells of Escherichia coli. J. Bacteriol. 2000, 182, 4337–4342. [Google Scholar] [CrossRef] [PubMed]
- Jens, M.; Philippe, E.; Ima Avalos, V.; Philip, K.; Viola, V. Bacterial filamentation accelerates colonization of adhesive spots embedded in biopassive surfaces. New J. Phys. 2013, 15, 125016. [Google Scholar]
- Lederer, F.L.; Gunther, T.J.; Flemming, K.; Raff, J.; Fahmy, K.; Springer, A.; Pollmann, K. Heterologous expression of the surface-layer-like protein SllB induces the formation of long filaments of Escherichia coli consisting of protein-stabilized outer membrane. Microbiology 2010, 156 Pt 12, 3584–3595. [Google Scholar] [CrossRef] [PubMed]
- Lederer, F.L.; Günther, T.J.; Weinert, U.; Raff, J.; Pollmann, K. Development of functionalised polyelectrolyte capsules using filamentous Escherichia coli cells. Microbial. Cell Factories 2012, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, S.; DiLuzio, W.R.; Weibel, D.B.; Whitesides, G.M. Controlling the Shape of Filamentous Cells of Escherichia coli. Nano Lett. 2005, 5, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Möller, J.; Emge, P.; Vizcarra, I.A.; Kollmannsberger, P.; Vogel, V. Bacterial filamentation accelerates colonization of adhesive spots embedded in biopassive surfaces. New J. Phys. 2013, 15. [Google Scholar] [CrossRef]
- Hong, Y.; Brown, D.G. Variation in Bacterial ATP Level and Proton Motive Force Due to Adhesion to a Solid Surface. Appl. Environ. Microbiol. 2009, 75, 2346–2353. [Google Scholar] [CrossRef] [PubMed]
- Albert, L.S.; Brown, D.G. Variation in bacterial ATP concentration during rapid changes in extracellular pH and implications for the activity of attached bacteria. Colloids Surf. B Biointerfaces 2015, 132, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Hol, F.J.H.; Dekker, C. Zooming in to see the bigger picture: Microfluidic and nanofabrication tools to study bacteria. Science 2014, 346, 438. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.C. The future is now: single-cell genomics of bacteria and archaea. Fems Microbiol. Rev. 2012, 37, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Wessel, A.K.; Hmelo, L.; Parsek, M.R.; Whiteley, M. Going local: Technologies for exploring bacterial microenvironments. Nat. Rev. Microbiol. 2013, 11, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, J.R.; Belkin, S. Where microbiology meets microengineering: Design and applications of reporter bacteria. Nat. Rev. Microbiol. 2010, 8, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Rosa, J.C.; Neto, A.B.; Hokka, C.O.; Badino, A.C. Influence of dissolved oxygen and shear conditions on clavulanic acid production by Streptomyces clavuligerus. Bioprocess Biosyst. Eng. 2005, 27, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Tsao, C.-.Y.; Wu, H.C.; Quan, D.N.; Payne, G.F.; Rubloff, G.W.; Bentley, W.E. Distal modulation of bacterial cell-cell signalling in a synthetic ecosystem using partitioned microfluidics. Lab Chip 2015, 15, 1842–1851. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, R.; Guasto, J.S.; Stocker, R. Bacterial transport suppressed by fluid shear. Nat. Phys. 2014, 10, 212–217. [Google Scholar] [CrossRef]
- Stroka, K.M.; Gu, Z.; Sun, S.X.; Konstantopoulos, K. Bioengineering paradigms for cell migration in confined microenvironments. Curr. Opin. Cell Biol. 2014, 30, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Guasto, J.S.; Rusconi, R.; Stocker, R. Fluid Mechanics of Planktonic Microorganisms. Annu. Rev. Fluid Mech. 2012, 44, 373–400. [Google Scholar] [CrossRef]
- Sipos, O.; Nagy, K.; Galajda, P. Patterns of Collective Bacterial Motion in Microfluidic Devices. Chem. Biochem. Eng. Q. 2014, 28, 233–240. [Google Scholar] [CrossRef]
- Karimi, A.; Karig, D.; Kumar, A.; Ardekani, A.M. Interplay of physical mechanisms and biofilm processes: Review of microfluidic methods. Lab Chip 2015, 15, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, R.; Lecuyer, S.; Autrusson, N.; Guglielmini, L.; Stone, H.A. Secondary Flow as a Mechanism for the Formation of Biofilm Streamers. Biophys. J. 2011, 100, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, Y.D.N.; Vogeleer, P.; Jacques, M.; Harel, J. High-Throughput Microfluidic Method to Study Biofilm Formation and Host-Pathogen Interactions in Pathogenic Escherichia coli. Appl. Environ. Microbiol. 2015, 81, 2827–2840. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, H.D.; Chung, S. Microfluidic approaches to bacterial biofilm formation. Molecules 2012, 17, 9818–9834. [Google Scholar] [CrossRef] [PubMed]
- Usami, S.; Chen, H.-.H.; Zhao, Y.; Chien, S.; Skalak, R. Design and construction of a linear shear stress flow chamber. Ann. Biomed. Eng. 1993, 21, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Kaya, T.; Koser, H. Direct upstream motility in Escherichia coli. Biophys. J. 2012, 102, 1514–1523. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; Kalkanci, O.; McMurry, J.L.; Koser, H. Hydrodynamic surface interactions enable Escherichia coli to seek efficient routes to swim upstream. Phys. Rev. Lett. 2007, 98, 068101. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. Sticky situations: Key components that control bacterial surface attachment. J. Bacteriol. 2012, 194, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Yakovenko, O.; Sharma, S.; Forero, M.; Tchesnokova, V.; Aprikian, P.; Kidd, B.; Mach, A.; Vogel, V.; Sokurenko, E.; Thomas, W.E. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J. Biol. Chem. 2008, 283, 11596–11605. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.J.; Yamashita, K.; Kobata, A. Structural study of the carbohydrate moiety of bovine pancreatic ribonuclease B. J. Biochem. 1980, 88, 51–58. [Google Scholar] [PubMed]
- Sokurenko, E.V.; Chesnokova, V.; Doyle, R.J.; Hasty, D.L. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J. Biol. Chem. 1997, 272, 17880–17886. [Google Scholar] [CrossRef] [PubMed]
- Wellens, A.; Garofalo, C.; Nguyen, H.; van Gerven, N.; Slättegård, R.; Hernalsteens, J.P.; Wyns, L.; Oscarson, S.; de Greve, H.; Hultgren, S. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS ONE 2008, 3, e2040. [Google Scholar] [CrossRef]
- Otto, K.; Norbeck, J.; Larsson, T.; Karlsson, K.A.; Hermansson, M. Adhesion of type 1-fimbriated Escherichia coli to abiotic surfaces leads to altered composition of outer membrane proteins. J. Bacteriol. 2001, 183, 2445–2453. [Google Scholar] [CrossRef] [PubMed]
- Otto, K.; Elwing, H.; Hermansson, M. The role of type 1 fimbriae in adhesion of Escherichia coli to hydrophilic and hydrophobic surfaces. Colloids Surf. B 1999, 15, 99–111. [Google Scholar] [CrossRef]
- Mulvey, M.A.; Lopez-Boado, Y.S.; Wilson, C.L.; Roth, R.; Parks, W.C.; Heuser, J.; Hultgren, S.J. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 1998, 282, 1494–1497. [Google Scholar] [CrossRef] [PubMed]
- Otto, K.; Silhavy, T.J. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 2287–2292. [Google Scholar] [CrossRef] [PubMed]
- Raivio, T.L.; Silhavy, T.J. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 2001, 55, 591–624. [Google Scholar] [CrossRef] [PubMed]
- Wickens, H.J.; Pinney, R.J.; Mason, D.J.; Gant, V.A. Flow cytometric investigation of filamentation, membrane patency, and membrane potential in Escherichia coli following ciprofloxacin exposure. Antimicrob. Agents Chemother. 2000, 44, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.L.; Hurley, M.M.; Jahnke, J.P.; Stratis-Cullum, D.N. Functional and Selective Bacterial Interfaces Using Cross-Scaffold Gold Binding Peptides. JOM 2015, 67, 2483–2493. [Google Scholar] [CrossRef]
- Goldstein, R.E.; Goriely, A. Dynamic buckling of morphoelastic filaments. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2006, 74 Pt 1, 010901. [Google Scholar] [CrossRef] [PubMed]
- Moulton, D.E.; Lessinnes, T.; Goriely, A. Morphoelastic rods. Part I: A single growing elastic rod. J. Mech. Phys. Solids 2013, 61, 398–427. [Google Scholar] [CrossRef]
- Honda, R.; Wakita, J.-I.; Katori, M. Self-Elongation with Sequential Folding of a Filament of Bacterial Cells. J. Phys. Soc. Jpn. 2015, 84, 114002. [Google Scholar] [CrossRef]
- El-Hajj, Z.W.; Newman, E.B. An Escherichia coli mutant that makes exceptionally long cells. J. Bacteriol. 2015, 197, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Klemm, P.; Jorgensen, B.J.; van Die, I.; de Ree, H.; Bergmans, H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol. Gen. Genet. 1985, 199, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Blomfield, I.C.; McClain, M.S.; Eisenstein, B.I. Type 1 fimbriae mutants of Escherichia coli K12: Characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 1991, 5, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Terrell, J.L.; Dong, H.; Holthoff, E.L.; Small, M.C.; Sarkes, D.A.; Hurley, M.M.; Stratis-Cullum, D.N. Investigation of engineered bacterial adhesins for opportunity to interface cells with abiotic materials. Proc. SPIE 9863 Smart Biomed. Physiol. Sens. Technol. XIII 2016, 986308. [Google Scholar] [CrossRef]
- Cheng, X.H.; Irimia, D.; Dixon, M.; Sekine, K.; Demirci, U.; Zamir, L.; Tompkins, R.G.; Rodriguez, W.; Toner, M. A microfluidic device for practical label-free CD4+T cell counting of HIV-infected subjects. Lab Chip 2007, 7, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the E. coli strains used in this study are available from the authors.





© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahnke, J.P.; Terrell, J.L.; Smith, A.M.; Cheng, X.; Stratis-Cullum, D.N. Influences of Adhesion Variability on the “Living” Dynamics of Filamentous Bacteria in Microfluidic Channels. Molecules 2016, 21, 985. https://doi.org/10.3390/molecules21080985
Jahnke JP, Terrell JL, Smith AM, Cheng X, Stratis-Cullum DN. Influences of Adhesion Variability on the “Living” Dynamics of Filamentous Bacteria in Microfluidic Channels. Molecules. 2016; 21(8):985. https://doi.org/10.3390/molecules21080985
Chicago/Turabian StyleJahnke, Justin P., Jessica L. Terrell, Austin M. Smith, Xuanhong Cheng, and Dimitra N. Stratis-Cullum. 2016. "Influences of Adhesion Variability on the “Living” Dynamics of Filamentous Bacteria in Microfluidic Channels" Molecules 21, no. 8: 985. https://doi.org/10.3390/molecules21080985
APA StyleJahnke, J. P., Terrell, J. L., Smith, A. M., Cheng, X., & Stratis-Cullum, D. N. (2016). Influences of Adhesion Variability on the “Living” Dynamics of Filamentous Bacteria in Microfluidic Channels. Molecules, 21(8), 985. https://doi.org/10.3390/molecules21080985