Synthesis and Molecular Modeling Studies of N′-Hydroxyindazolecarboximidamides as Novel Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
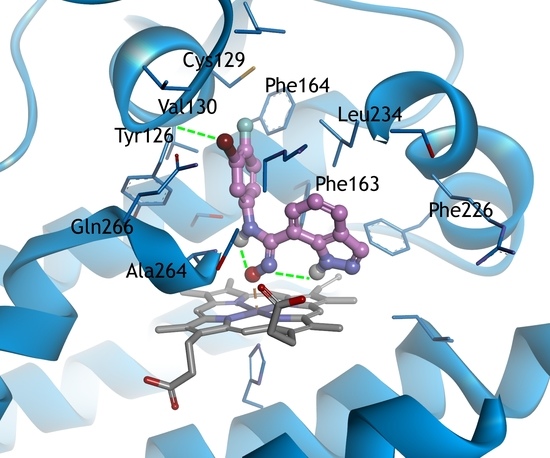
2.3. Molecular Docking Study
3. Materials and Methods
3.1. Chemistry
3.1.1. General Chemistry
3.1.2. General Procedure for the Preparation of Compounds
Synthesis of 1H-Indazol-7-amine 3
Synthesis of 7-Iodo-1H-indazole 4
Synthesis of 1H-Indazole-7-carbonitrile 5
Synthesis of (Z)-N′-Hydroxy-1H-indazole-7-carboximidamide 6
Synthesis of (E)-N-Hydroxy-1H-indazole-7-carbimidoyl chloride 7
General Procedure for the Synthesis of 8, 12 and 16
Synthesis of N-Hydroxy-1-methyl-1H-indazole-7-carbimidoyl chloride (11)
3.2. Biology
3.2.1. Generation of Human Ido1 Gene Expressing Hek293 Recombinant Cells
3.2.2. Cell Based Assay for Analysis of Anti-Ido1 Activity of Compounds by Determination of Tryptophan and Kynurenine Using an LC-MS System
3.2.3. hIDO1 Enzymatic Assay
3.3. Docking Study
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lob, S.; Konigsrainer, A.; Rammensee, H.G.; Opelz, G.; Terness, P. Inhibitors of indoleamine-2,3-dioxygenase for cancer therapy: Can we see the wood for the trees? Nat. Rev. Cancer 2009, 9, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Mellor, A.L. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J. Clin. Investig. 2007, 117, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Hwu, P.; Du, M.X.; Lapointe, R.; Do, M.; Taylor, M.W.; Young, H.A. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J. Immunol. 2000, 164, 3596–3599. [Google Scholar] [CrossRef] [PubMed]
- Curti, A.; Trabanelli, S.; Salvestrini, V.; Baccarani, M.; Lemoli, R.M. The role of indoleamine 2,3-dioxygenase in the induction of immune tolerance: Focus on hematology. Blood 2009, 113, 2394–2401. [Google Scholar] [CrossRef] [PubMed]
- Palafox, D.; Llorente, L.; Alberu, J.; Torres-Machorro, A.; Camorlinga, N.; Rodriguez, C.; Granados, J. The role of indoleamine 2,3 dioxygenase in the induction of immune tolerance in organ transplantation. Transplant. Rev. 2010, 24, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Uyttenhove, C.; Pilotte, L.; Theate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Brandacher, G.; Perathoner, A.; Ladurner, R.; Schneeberger, S.; Obrist, P.; Winkler, C.; Werner, E.R.; Werner-Felmayer, G.; Weiss, H.G.; Gobel, G.; et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin. Cancer Res. 2006, 12, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Ferdinande, L.; Decaestecker, C.; Verset, L.; Mathieu, A.; Moles Lopez, X.; Negulescu, A.M.; Van Maerken, T.; Salmon, I.; Cuvelier, C.A.; Demetter, P. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. Br. J. Cancer. 2012, 106, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shen, Z.; Wang, Z.; Wang, X.; Zhang, H.; Qin, J.; Qin, X.; Xu, J.; Sun, Y. Increased expression of IDO associates with poor postoperative clinical outcome of patients with gastric adenocarcinoma. Sci. Rep. 2016, 6, 21319. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C. IDO inhibitors move center stage in immuno-oncology. Nat. Biotechnol. 2015, 33, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Rohrig, U.F.; Majjigapu, S.R.; Vogel, P.; Zoete, V.; Michielin, O. Challenges in the Discovery of Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors. J. Med. Chem. 2015, 58, 9421–9437. [Google Scholar] [CrossRef] [PubMed]
- Weinmann, H. Cancer Immunotherapy: Selected Targets and Small-Molecule Modulators. ChemMedChem 2016, 11, 450–466. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.W.; Douty, B.; Wayland, B.; Bower, M.; Liu, X.; Leffet, L.; Wang, Q.; Bowman, K.J.; Hansbury, M.J.; Liu, C.; et al. Discovery of potent competitive inhibitors of indoleamine 2,3-dioxygenase with in vivo pharmacodynamic activity and efficacy in a mouse melanoma model. J. Med. Chem. 2009, 52, 7364–7367. [Google Scholar] [CrossRef] [PubMed]
- Mautino, M.; Kumar, S.; Waldo, J.; Jaipuri, F.; Kesharwani, T. Imidazole Derivatives as IDO Inhibitors. Patent WO2011/056652, 21 May 2011. [Google Scholar]
- Kumar, S.; Waldo, J.; Jaipuri, F.; Mautino, M. Tricyclic Compoundsasinhibitorsofimmunosuppression Mediated by Tryptophan Metabolization. Patent WO2014/159248, 2 October 2014. [Google Scholar]
- Peng, Y.; Ueng, S.; Tseng, C.; Hung, M.; Song, J.; Wu, J.; Liao, F.; Fan, Y.; Wu, M.; Hsiao, W.; et al. Important Hydrogen Bond Networks in Indoleamine 2,3-dioxygenase 1 (IDO1) Inhibitor Design revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1. J. Med. Chem. 2016, 59, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.W.; Sparks, R.; Polam, P.; Modi, D.; Douty, B.; Wayland, B.; Glass, B.; Takvorian, A.; Glenn, J.; Zhu, W.; et al. INCB24360 (Epacadostat), a Highly Potent and Selective Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitor for Immuno-oncology. ACS Med. Chem. Lett. 2017, 8, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, T.; Liu, J.; Ding, K.; Xu, J. Structural insights into the binding mechanism of IDO1 with hydroxylamidine based inhibitor INCB14943. Biochem. Biophys. Res. Commun. 2017, 487, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Cottyn, B.; Acher, F.; Ramassamy, B.; Alvey, L.; Lepoivre, M.; Frapart, Y.; Stuehr, D.; Mansuy, D.; Boucher, J.L.; Vichard, D. Inhibitory effects of a series of 7-substituted-indazoles toward nitric oxide synthases: particular potency of 1H-indazole-7-carbonitrile. Bioorg. Med. Chem. 2008, 16, 5962–5973. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Si, P.; Wang, L.; Xu, Y.; Xu, X.; Zhu, J.; Jiang, H.; Li, W.; Chen, L.; Li, J. Design, Synthesis, and Biological Evaluation of Novel Nonsteroidal Farnesoid X Receptor (FXR) Antagonists: Molecular Basis of FXR Antagonism. ChemMedChem 2015, 10, 1184–1199. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Yang, J.; Park, S.; Jo, E.; Kim, H.Y.; Bae, Y.S.; Windisch, M.P. Micrococcin P1, a naturally occurring macrocyclic peptide inhibiting hepatitis C virus entry in a pan-genotypic manner. Antiviral Res. 2016, 132, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Schrodinger. Available online: http://www.schrodinger.com (accessed on 8 November 2017).
- RCSB PDB. Available online: http://www.rcsb.org/pdb (accessed on 8 November 2017).
- Biovia. Available online: http://www.biovia.com (accessed on 8 November 2017).
Sample Availability: Samples of the compounds 8, 12 and 16 are available from the authors. |





| No. | R1 | HEK293 IC50 (µM) | hIDO1 Inhibition at 20 µM (%) | |
|---|---|---|---|---|
| Tryptophan Depletion | Kynurenine Production | |||
| 8a | 3-Br-4-F | 1.76 | 2.13 | 81 |
| 8b | 3-Cl-4-F | 3.26 | 3.14 | 79 |
| 8c | 3-Cl | 4.95 | 4.89 | 77 |
| 8d | 4-F | >10 | >10 | n.t. |
| 8e | 2,5-diF | >10 | >10 | n.t. |
| 8f | 3-Ac | >10 | >10 | n.t. |
| 8g | 3-Me | >10 | >10 | n.t. |
| 8h | 4-Cl-3-Me | >10 | >10 | n.t. |
| 8i | 3-Cl-4-MeO | >10 | >10 | n.t. |
| 8j | H | >10 | >10 | n.t. |
| 8k | 4-Cl | >10 | >10 | n.t. |
| 8l | 4-MeO | >10 | >10 | n.t. |
| 8m | 4-Me | >10 | >10 | n.t. |
| 12 |  | >10 | >10 | 15 |
| 16 |  | 7.76 | 7.91 | n.t. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.-H.; Lee, J.-Y.; Jeong, J.; Kim, M.; Lee, K.W.; Jang, E.; Ahn, S.; Lee, C.H.; Hwang, J.Y. Synthesis and Molecular Modeling Studies of N′-Hydroxyindazolecarboximidamides as Novel Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors. Molecules 2017, 22, 1936. https://doi.org/10.3390/molecules22111936
Lee D-H, Lee J-Y, Jeong J, Kim M, Lee KW, Jang E, Ahn S, Lee CH, Hwang JY. Synthesis and Molecular Modeling Studies of N′-Hydroxyindazolecarboximidamides as Novel Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors. Molecules. 2017; 22(11):1936. https://doi.org/10.3390/molecules22111936
Chicago/Turabian StyleLee, Dong-Ho, Joo-Youn Lee, Jieun Jeong, Miok Kim, Kyung Won Lee, Eunseo Jang, Sunjoo Ahn, Chang Hoon Lee, and Jong Yeon Hwang. 2017. "Synthesis and Molecular Modeling Studies of N′-Hydroxyindazolecarboximidamides as Novel Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors" Molecules 22, no. 11: 1936. https://doi.org/10.3390/molecules22111936
APA StyleLee, D. -H., Lee, J. -Y., Jeong, J., Kim, M., Lee, K. W., Jang, E., Ahn, S., Lee, C. H., & Hwang, J. Y. (2017). Synthesis and Molecular Modeling Studies of N′-Hydroxyindazolecarboximidamides as Novel Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors. Molecules, 22(11), 1936. https://doi.org/10.3390/molecules22111936