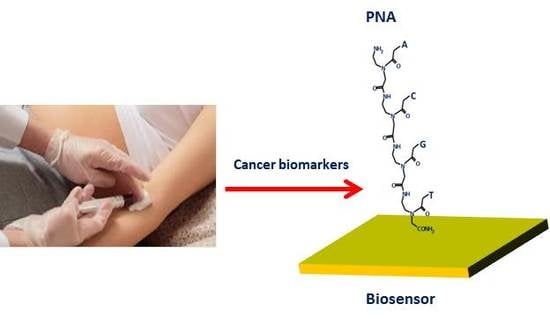
Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis
Abstract
:1. Introduction
2. PNA-Based Biosensors
2.1. PNA-Based Biosensors for RNA Detection
2.2. PNA-Based Biosensors for DNA Detection
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dancey, J.E.; Bedard, P.L.; Onetto, N.; Hudson, T.J. The genetic basis for cancer treatment decisions. Cell 2012, 148, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Diamandis, M.; White, N.M.; Yousef, G.M. Personalized medicine: marking a new epoch in cancer patient management. Mol. Cancer Res. 2010, 8, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Nguyen, Q.T. Molecular imaging for cancer diagnosis and surgery. Adv. Drug Deliv. Rev. 2014, 66, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. Exosomes/microvesicles: Mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol. 2011, 33, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Luna Coronell, J.A.; Syed, P.; Sergelen, K.; Gyurján, I.; Weinhäusel, A. The current status of cancer biomarker research using tumour-associated antigens for minimal invasive and early cancer diagnostics. J. Proteom. 2012, 76, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Kelley, S.O. Advancing ultrasensitive molecular and cellular analysis methods to speed and simplify the diagnosis of disease. Acc. Chem. Res. 2017, 50, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Bellassai, N.; Spoto, G. Biosensors for liquid biopsy: Circulating nucleic acids to diagnose and treat cancer. Anal. Bioanal. Chem. 2016, 408, 7255–7264. [Google Scholar] [CrossRef] [PubMed]
- Ranjana, R.; Esimbekova, E.N.; Kratasyuka, V.A. Rapid biosensing tools for cancer biomarkers. Biosens. Bioelectron. 2017, 87, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Hamburg, M.A.; Collins, F.S. The path to personalized medicine. N. Engl. J. Med. 2010, 363, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; David, B.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Wang, W. From cleanroom to desktop: Emerging micro-nanofabrication technology for biomedical applications. Ann. Biomed. Eng. 2011, 39, 600–620. [Google Scholar] [CrossRef] [PubMed]
- Krabbenborg, S.O.; Nicosia, C.; Chen, P.; Huskens, J. Reactivity mapping with electrochemical gradients for monitoring reactivity at surfaces in space and time. Nat. Commun. 2013, 4, 1667. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; D’Agata, R.; Spoto, G. Functionalized gold nanoparticles for ultrasensitive DNA detection. Anal. Bioanal. Chem. 2012, 402, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cushing, S.K.; Wu, N. Plasmon-enhanced optical sensors: A review. Analyst 2015, 140, 386–406. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Palladino, P.; Spoto, G. Streptavidin-coated gold nanoparticles: Critical role of oligonucleotides on stability and fractal aggregation. Beilstein J. Nanotechnol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Swierczewska, M.; Liu, G.; Lee, S.; Chen, X. High-sensitivity nanosensors for biomarker detection. Chem. Soc. Rev. 2012, 41, 2641–2655. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M. Cancer nanotechnology: Opportunities and challenges. Nat. Rev. Cancer 2005, 5, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Kelley, S.O.; Mirkin, C.A.; Walt, D.R.; Ismagilov, R.F.; Toner, M.; Sargent, E.H. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat. Nanotechnol. 2014, 9, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Bala, A.; Goorski, Ł. Application of nucleic acid analogues as receptor layers for biosensors. Anal. Methods 2016, 8, 236–244. [Google Scholar] [CrossRef]
- D’Agata, R.; Spoto, G. Artificial DNA and surface plasmon resonance. Artif. DNA PNA XNA 2012, 3, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P. Peptide nucleic acids (PNA) in chemical biology and drug discovery. Chem. Biodivers. 2010, 7, 786–804. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Koshkin, A.A.; Wengel, J.; Nielsen, P. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 0, 455–456. [Google Scholar] [CrossRef]
- Spoto, G.; Corradini, R. Detection of Non-Amplified Genomic DNA; Springer: Dordrecht, The Netherlands, 2012; p. 315. [Google Scholar]
- Lundin, K.; Good, L.; Strömberg, R.; Gräslund, A.; Smith, C.I.E. Biological activity and biotechnological aspects of peptide nucleic acid. Adv. Genet. 2006, 56, 1–51. [Google Scholar] [PubMed]
- Briones, C.; Moreno, M. Applications of peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) in biosensor development. Anal. Bioanal. Chem. 2012, 402, 3071–3089. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.R.; Lee, J.; Yeo, J.; Na, H.K.; Kim, Y.K.; Jang, H.; Lee, J.H.; Han, S.W.; Lee, Y.; Kim, V.N.; et al. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano 2013, 23, 5882–5891. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Han, J.; Xue, P.; Xu, R.; Kang, Y. Nano metal-organic framework (NMOF)-based strategies for multiplexed microRNA detection in solution and living cancer cells. Nanoscale 2015, 7, 1753–1759. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, Q.; Ju, H. A peptide nucleic acid-functionalized carbon nitride nanosheet as a probe for in situ monitoring of intracellular microRNA. Analyst 2015, 140, 4245–4252. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, U.H.; Alagappan, P.; Liedberg, B. Naked eye detection of lung cancer associated miRNA by paper based biosensing platform. Anal. Chem. 2013, 85, 820–824. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, A.; Cheema, J.A.; Rajwar, D.; Ammanath, G.; Xiaohu, L.; Koon, L.S.; Yi, W.; Yildizd, U.H.; Liedberg, B. Polythiophene derivative on quartz resonators for miRNA capture and assay. Analyst 2015, 140, 7912–7917. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Xu, Y.; Luo, Y.; Zhu, L.; Zhang, Y.; Huang, K.; Xu, W. Specific and relative detection of urinary microRNA signatures in bladder cancer for point-of-care diagnostics. Chem. Commun. 2017, 53, 4222–4225. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Batistuti, M.R.; Miodek, A.; Zhurauski, P.; Mulato, M.; Lindsay, M.A.; Estrela, P. Highly sensitive dual mode electrochemical platform for microRNA detection. Sci. Rep. 2016, 6, 36719. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Shen, W.; Ren, Y.; Gao, Z. A highly sensitive microRNA biosensor based on hybridized microRNA-guided deposition of polyaniline. Biosens. Bioelectron. 2014, 60, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Huang, L.; Zhang, H.; Sun, Z.; Zhang, Z.; Zhang, G.J. Gold nanoparticles-decorated graphene field-effect transistor biosensor for femtomolar MicroRNA detection. Biosens. Bioelectron. 2015, 74, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.H.; Sim, S.J. Nanoplasmonic biosensor: Detection and amplification of dual bio-signatures of circulating tumor DNA. Biosens. Bioelectron. 2015, 67, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Jampasa, S.; Wonsawat, W.; Rodthongkum, N.; Siangproh, W.; Yanatatsaneejit, P.; Tirayut, V.; Chailapakul, O. Electrochemical detection of human papilloma virus DNA type16 using a pyrrolidinyl peptid e nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014, 54, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Henry, C.S.; Vilaivan, T.; Chailapakul, O. Electrochemical paper-based peptide nucleic acid biosensor for detecting human papillomavirus. Anal. Chim. Acta 2017, 952, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Da, J.; Ivanov, I.; Montermini, L.; Rak, J.; Sargent, E.H.; Kelley, S.O. An electrochemical clamp assay for direct, rapid analysis of circulating nucleic acids in serum. Nat. Chem. 2015, 7, 569–575. [Google Scholar]
- Das, J.; Ivanov, I.; Sargent, E.H.; Kelley, S.O. DNA clutch probes for circulating tumor DNA analysis. J. Am. Chem. Soc. 2016, 138, 11009–11016. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M. An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1999, 1, 89–104. [Google Scholar] [PubMed]
- Uhlmann, E.; Peyman, A.; Breipohl, G.; Will, D.W. PNA: Synthetic polyamide nucleic acids with unusual binding properties. Angew. Chem. Int. Ed. 1998, 37, 2796–2823. [Google Scholar] [CrossRef]
- Park, H.; Germini, A.; Sforza, S.; Corradini, R.; Marchelli, R.; Knoll, W. Kinetic and affinity analyses of hybridization reactions between peptide nucleic acid probes and DNA targets using surface plasmon field-enhanced fluorescence spectroscopy. Biointerphases 2006, 1, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Ratilainen, T.; Holmen, A.; Tuite, E.; Nielsen, P.E.; Norden, B. Thermodynamics of sequence-specific binding of PNA to DNA. Biochemistry 2000, 39, 7781–7791. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Awasthi, S.K. Versatility of peptide nucleic acids (PNAs): Role in chemical biology, drug discovery, and origins of life. Chem. Biol. Drug Des. 2017, 89, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Gambari, R. Peptide nucleic acids: A review on recent patents and technology transfer. Expert Opin. Ther. Pat. 2014, 24, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Oh, B.K.; Choi, J.W. Application of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochemistry 2010, 79, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Demidov, V.V.; Protozanova, E.; Izvolsky, K.I.; Price, C.; Nielsen, P.E.; Frank-Kamenetskii, M.D. Kinetics and mechanism of the DNA double helix invasion by pseudocomplementary peptide nucleic acids. Proc. Natl. Acad. Sci. USA 2002, 99, 5953–5958. [Google Scholar] [CrossRef] [PubMed]
- Hanvey, J.C.; Peffer, N.J.; Bisi, J.E.; Thomson, S.A.; Cadilla, R.; Josey, J.A.; Ricca, D.J.; Hassman, C.F.; Bonham, M.A.; Au, K.G. Antisense and antigene properties of peptide nucleic acids. Science 1992, 258, 1481–1485. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, E.; Brognara, E.; Borgatti, M.; Lampronti, I.; Finotti, A.; Bianchi, N.; Sforza, S.; Tedeschi, T.; Manicardi, A.; Marchelli, R.; et al. miRNA therapeutics: Delivery and biological activity of peptide nucleic acids targeting miRNAs. Epigenomics 2011, 3, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Koppelhus, U.; Awasthi, S.K.; Zachar, V.; Uffe Holst, H.; Ebbesen, P.; Nielsen, P.E. Cell-dependent differential cellular uptake of PNA, peptides, and PNA-peptide conjugates. Antisense Nucleic Acid Drug Dev. 2002, 12, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Corradini, R.; Sforza, S.; Tedeschi, T.; Totsingan, F.; Manicardi, A.; Marchelli, R. Peptide nucleic acids with a structurally biased backbone. Updated review and emerging challenges. Curr. Top. Med. Chem. 2011, 11, 1535–1554. [Google Scholar] [CrossRef] [PubMed]
- Manicardi, A.; Rozzi, A.; Korom, S.; Corradini, R. Building on the peptide nucleic acid (PNA) scaffold: A biomolecular engineering approach. J. Supramol. Chem. 2017, 1–12. [Google Scholar] [CrossRef]
- Ørum, H. PCR clamping. In Peptide Nucleic Acids, Protocols and Applications; Nielsen, P.E., Ed.; Horizon Bioscience: Wymondham, UK, 2004; pp. 175–185. [Google Scholar]
- Taback, B.; Bilchik, A.J.; Saha, S.; Nakayama, T.; Wiese, D.A.; Turner, R.R.; Kuo, C.T.; Hoon, D.S.B. Peptide nucleic acid clamp PCR: A novel K-ras mutation detection assay for colorectal cancer micrometastases in lymph nodes. Int. J. Cancer 2004, 111, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Miyano, S.; Hanazawa, K.; Kitabatake, T.; Fujisawa, M.; Kojima, K. Detecting KRAS mutations in peripheral blood of colorectal cancer patients by peptide nucleic acid clamp PCR. Exp. Ther. Med. 2012, 4, 790–794. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Liu, M.; Yu, L.; Cai, G.; Chen, Q.; Wu, R.; Wang, F.; Zhang, B.; Jiang, T.; Fu, W. Construction of a novel peptide nucleic acid piezoelectric gene sensor microarray detection system. J. Nanosci. Nanotechnol. 2005, 5, 1266–1272. [Google Scholar] [CrossRef] [PubMed]
- Ananthanawat, C.; Vilaivan, T.; Hoven, V.P. Synthesis and immobilization of thiolated pyrrolidinyl peptide nucleic acids on gold-coated piezoelectric quartz crystals for the detection of DNA hybridization. Sens. Actuator B 2009, 137, 215–221. [Google Scholar] [CrossRef]
- D’Agata, R.; Corradini, R.; Grasso, G.; Marchelli, R.; Spoto, G. Ultrasensitive detection of DNA by PNA and nanoparticle-enhanced surface plasmon resonance imaging. ChemBioChem 2008, 9, 2067–2070. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Corradini, R.; Ferretti, C.; Zanoli, L.; Gatti, M.; Marchelli, R.; Spoto, G. Ultrasensitive detection of non-amplified genomic DNA by nanoparticle-enhanced surface plasmon resonance imaging. Biosens. Bioelectron. 2010, 25, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Lao, A.I.K.; Su, X.; Aung, K.M.M. SPR study of DNA hybridization with DNA and PNA probes under stringent conditions. Biosens. Bioelectron. 2009, 24, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, A.; Wasserberg, D.; Posthuma-Trumpie, G.A.; Subramaniam, V.; van Amerongen, A.; Corradini, R.; Tedeschi, T.; Sforza, S.; Reinhoudt, D.N.; Marchelli, R.; et al. Patterning of peptide nucleic acids using reactive microcontact printing. Langmuir 2011, 27, 1536–1542. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yang, F.; Li, W.; Zhao, W.; Nie, K.; Dong, B.; Liu, Z. A review: Fabrications, detections and applications of peptide nucleic acids (PNAs) microarray. Biosens. Bioelectron. 2015, 66, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.W.; Choi, S.W.; You, B.H. Incredible RNA: Dual functions of coding and noncoding. Mol. Cells 2016, 39, 367–374. [Google Scholar] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Mercado, M.; Manterola, L.; Larrea, E.; Goicoechea, I.; Arestin, M.; Armesto, M.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of non-invasive cancer biomarkers: Concepts and controversies of non-coding and coding RNA in body fluids. J. Cell. Mol. Med. 2015, 19, 2307–2323. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hrdlickova, B.; de Almeida, R.C.; Borek, Z.; Withoff, S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim. Biophys. Acta 2014, 1842, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Nana-Sinkam, S.P.; Croce, C.M. Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther. 2013, 93, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Mayr, M. MicroRNAs in vascular and metabolic disease. Circ. Res. 2012, 110, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Qinghua, C. An analysis of human microRNA and disease associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Li, W.; Li, Y.; Tan, C.; Li, J.; Jin, Y.; Ruan, K. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005, 33, e17. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, M.; Schmidt, S.; Benes, V.; Noerholm, M.; Kulozik, A.E.; Hentze, M.W.; Muckenthaler, M.U. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA 2006, 12, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Miura, K.; Iwao, H.; Yamanaka, S. Quantitative assessment of DNA microarrays-comparison with Northern blot analysis. Genomics 2001, 71, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Arden, E.K.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yao, B.; Huang, H.; Wang, Z.; Sun, C.; Fan, Y.; Chang, Q.; Li, S.; Wang, X.; Xi, J. Real-time polymerase chain reaction microRNA detection based on enzymatic stem-loop probes ligation. Anal. Chem. 2009, 81, 5446–5451. [Google Scholar] [CrossRef] [PubMed]
- Driskell, J.D.; Seto, A.G.; Jones, L.P.; Jokela, S.; Dluhy, R.A.; Zhao, Y.P.; Tripp, R.A. Rapid microRNA (miRNA) detection and classification via surface-enhanced Raman spectroscopy (SERS). Biosens. Bioelectron. 2008, 24, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Muniz-Miranda, M.; Gellini, C.; Pagliai, M.; Innocenti, M.; Salvi, P.R.; Schettino, V. SERS and computational studies on microRNA chains adsorbed on silver surfaces. J. Phys. Chem. C 2010, 114, 13730–13735. [Google Scholar] [CrossRef]
- Cho, H.; Lee, B.; Liu, G.L.; Agarwal, A.; Lee, L.P. Label-free and highly sensitive biomolecular detection using SERS and electrokinetic preconcentration. Lab Chip 2009, 9, 3360–3363. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, M.C.; Zanoli, L.M.; D’Agata, R.; Finotti, A.; Gambari, R.; Spoto, G. Isothermal circular-strand-displacement polymerization of DNA and microRNA in digital microfluidic devices. Anal. Bioanal. Chem. 2015, 407, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef] [PubMed]
- Cissell, K.A.; Shrestha, S.; Deo, S.K. MicroRNA detection: Challenges for the analytical chemist. Anal. Chem. 2007, 79, 4754–4761. [Google Scholar] [CrossRef]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Graybill, R.M.; Bailey, R.C. Emerging biosensing approaches for microRNA analysis. Anal. Chem. 2016, 88, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Degliangeli, F.; Pompa, P.P.; Fiammengo, R. Nanotechnology-based strategies for the detection and quantification of microRNA. Chem. Eur. J. 2014, 20, 9476–9492. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Lyu, J.; Shi, J.; Yang, M. Graphene and graphene-like two-denominational materials based fluorescence resonance energy transfer (FRET) assays for biological applications. Biosens. Bioelectron. 2017, 89, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Eddaoudi, M.; O’Keeffe, M.; Yaghi, O.M. Design and synthesis of an exceptionally stable and highly porous metal-organic framework. Nature 1999, 402, 276–279. [Google Scholar]
- Zhu, X.; Zheng, H.Y.; Wei, X.F.; Lin, Z.Y.; Guo, L.H.; Qiu, B.; Chen, G.N. Metal–organic framework (MOF): A novel sensing platform for biomolecules. Chem. Commun. 2013, 49, 1276–1278. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, W.; Lei, J.; Xu, N.; Gao, F.; Ju, H. Fluorescence quenching of carbon nitride nanosheet through its interaction with DNA for versatile fluorescence sensing. Anal. Chem. 2013, 85, 12182–12188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xie, X.; Wang, H.; Zhang, J.; Pan, B.; Xie, Y. Enhanced photoresponsive ultrathin graphitic-phase C3N4 nanosheets for bioimaging. J. Am. Chem. Soc. 2013, 135, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Zhang, L.; Zhang, Y.; Lan, F.; Yan, M.; Yu, J. Nanomaterials-modified cellulose paper as a platform for biosensing applications. Nanoscale 2017, 9, 4366–4382. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; Spoto, G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, M.C.; Spoto, G. Integration of isothermal amplification methods in microfluidic devices: Recent advances. Biosens. Bioelectron. 2017, 90, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, Z.; Liu, C.; Cheng, Y. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew. Chem. 2010, 49, 5498–5501. [Google Scholar]
- Hamidi-Asl, E.; Palchetti, I.; Hasheminejad, E.; Mascini, M. A review on the electrochemical biosensors for determination of microRNAs. Talanta 2013, 115, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Wang, S.; Huang, L.; Ning, Y.; Zhang, Z.; Zhang, G.J. Ultrasensitive label-free detection of PNA–DNA hybridization by reduced graphene oxide field-effect transistor biosensor. ACS Nano 2014, 8, 2632–2638. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Abbott, J.; Qin, L.; Yeung, K.Y.; Song, Y.; Yoon, H.; Kong, J.; Ham, D. Electrophoretic and field-effect graphene for all-electrical DNA array technology. Nat. Commun. 2014, 5, 4866. [Google Scholar] [CrossRef] [PubMed]
- Ortmann, C.A.; Kent, D.G.; Nangalia, J.; Silber, Y.; Wedge, D.C.; Grinfeld, J.; Baxter, E.J.; Massie, C.E.; Papaemmanuil, E.; Menon, S.; et al. Effect of mutation order on myeloproliferative neoplasms. N. Engl. J. Med. 2015, 372, 1865–1866. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C. Real-time liquid biopsy in cancer patients: Fact or fiction? Cancer Res. 2013, 73, 6384–6388. [Google Scholar] [CrossRef] [PubMed]
- Lianidou, E.S. Circulating tumor cells—New challenges ahead. Clin. Chem. 2012, 58, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Kyung-A, H.; Junmoo, K.; Hogyeong, G.; Hyo-Il, J. Isolation and enrichment of circulating biomarkers for cancer screening, detection, and diagnostics. Analyst 2016, 141, 382–392. [Google Scholar]
- Thierry, A.R. A Targeted Q-PCR-based method for point mutation testing by analyzing circulating DNA for cancer management care. Methods Mol. Biol. 2016, 1392, 1–16. [Google Scholar] [PubMed]
- Yung, T.K.; Chan, K.C.; Mok, T.S.; Tong, J.; To, K.F.; Lo, Y.M. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin. Cancer Res. 2009, 15, 2076–2084. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra168. [Google Scholar] [CrossRef] [PubMed]
- Leary, R.J.; Sausen, M.; Kinde, I.; Papadopoulos, N.; Carpten, J.D.; Craig, D.; O’Shaughnessy, J.; Kinzler, K.W.; Parmigiani, G.; Vogelstein, B.; et al. Detection of chromosomal alterations in the circulation of cancer patients with whole genome sequencing. Sci. Transl. Med. 2012, 4, 162ra154. [Google Scholar] [CrossRef] [PubMed]
- Mauger, F.; Dulary, C.; Daviaud, C.; Deleuze, J.F.; Tost, J. Comprehensive evaluation of methods to isolate, quantify and characterize circulating cell-free DNA from small volumes of plasma. Anal. Bioanal. Chem. 2015, 407, 6873–6878. [Google Scholar] [CrossRef] [PubMed]
- Sau, T.K.; Rogach, A.L.; Jäckel, F.; Klar, T.A.; Feldmann, J. Properties and applications of colloidal nonspherical noble metal nanoparticles. Adv. Mater. 2010, 22, 1805–1825. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Gao, X.; Liu, D.; Chen, X. Gold nanoparticles for in vitro diagnostics. Chem Rev. 2015, 115, 10575–10636. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.M.; Hafner, J.H. Localized surface plasmon resonance sensors. Chem. Rev. 2011, 111, 3828–3857. [Google Scholar] [CrossRef] [PubMed]
- Devi, R.V.; Doble, M.; Verma, R.S. Nanomaterials for early detection of cancer biomarker with special emphasis on gold nanoparticles in immunoassays/sensors. Biosens. Bioelectron. 2015, 68, 688–698. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Spoto, G. Surface plasmon resonance imaging for nucleic acid detection. Anal. Bioanal. Chem. 2013, 405, 573–584. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Breveglieri, G.; Zanoli, L.M.; Borgatti, M.; Spoto, G.; Gambari, R. Direct detection of point mutations in nonamplified human genomic DNA. Anal Chem. 2011, 83, 8711–8717. [Google Scholar]
- Püschl, A.; Tedeschi, T.; Nielsen, P.E. Pyrrolidine PNA: A novel conformationally restricted PNA analogue. Org. Lett. 2000, 2, 4161–4163. [Google Scholar] [CrossRef] [PubMed]
- Green, B.J.; Saberi Safaei, T.; Mepham, A.; Labib, M.; Mohamadi, R.M.; Kelley, S.O. Beyond the capture of circulating tumor cells: Next-generation devices and materials. Angew. Chem. Int. Ed. 2016, 55, 1252–1265. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Itonaga, M.; Matsuzaki, I.; Warigaya, K.; Tamura, T.; Shimizu, Y.; Fujimoto, M.; Kojima, F.; Ichinose, M.; Murata, S. Novel methodology for rapid detection of KRAS mutation using PNA-LNA mediated loop-mediated isothermal amplification. PLoS ONE 2016, 11, e0151654. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Choi, Y.D.; Oh, I.J.; Kim, K.S.; Choi, H.; Chang, J.; Shin, H.J.; Park, C.K.; Kim, Y.C. Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non-small cell lung cancer. Cancer Res. Treat. 2015, 47, 661–669. [Google Scholar] [CrossRef] [PubMed]


| Target | Transduced Signal | LOD | Detection in Human Serum or Plasma | Reference |
|---|---|---|---|---|
| miR-21, miR-96 and miR-125b | Fluorescence | <1 pM | No | [31] |
| miR-21, miR-96 and miR-125b | Fluorescence | 10 pM | No | [32] |
| miR-18a | Fluorescence | - | No | [33] |
| miR-21 | Fluorescence | 10 nM | No | [34] |
| miR-21 | QCM | 400 pM | Yes | [35] |
| miR-126, miR-182 and miR-152 | Optical (Lateral flow test strip) | 0.6 fM | No | [36] |
| miR-145 | Electrochemical (Impedimetric and square-wave voltammetry) | 0.37 fM | No | [37] |
| miR let-7a, let-7b, let-7c | Electrochemical (Impedimetric) | 0.50 fM | No | [38] |
| miR let-7b, let-7c and miR 21 | Electric (Graphene field-effect transistor) | <10 fM | Yes | [39] |
| E542K, E545K, methylation in PIK3CA gene | Optical (Localized surface plasmon resonance) | 50 fM | Yes | [40] |
| HPV type 16 DNA, HPV types 18, 31 and 33 | Electrochemical (Square-wave voltammetry) | 4nM | No | [41] |
| DNA HPV type 16, type 18, type 31, and type 33 | Electrochemical (Impedimetric) | 2.3 nM | No | [42] |
| BRAF and KRAS DNA mutations | Electrochemical | 1 fg μL−1 | Yes | [43] |
| BRAF and KRAS DNA mutations | Electrochemical | 1 fg μL−1 | Yes | [44] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Agata, R.; Giuffrida, M.C.; Spoto, G. Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis. Molecules 2017, 22, 1951. https://doi.org/10.3390/molecules22111951
D’Agata R, Giuffrida MC, Spoto G. Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis. Molecules. 2017; 22(11):1951. https://doi.org/10.3390/molecules22111951
Chicago/Turabian StyleD’Agata, Roberta, Maria Chiara Giuffrida, and Giuseppe Spoto. 2017. "Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis" Molecules 22, no. 11: 1951. https://doi.org/10.3390/molecules22111951
APA StyleD’Agata, R., Giuffrida, M. C., & Spoto, G. (2017). Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis. Molecules, 22(11), 1951. https://doi.org/10.3390/molecules22111951