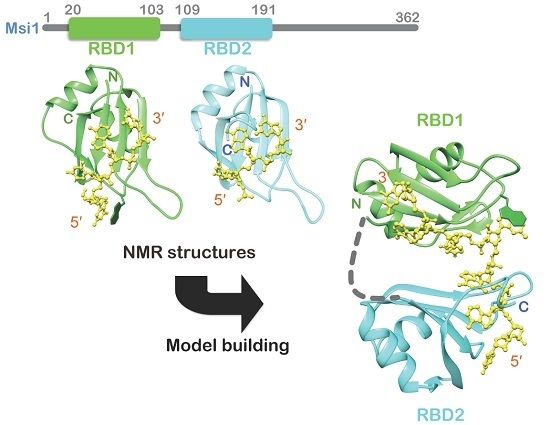
Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Abstract
:1. Introduction
2. Results
2.1. Structure Determination of Msi1 RBD2(109–191) in the Free Form
2.2. NMR Titration Experiments on Msi1(109–200) with r(GUAGU)
2.3. Structure Determination of the Msi1 RBD2(109–191):r(GUAGU) Complex
2.4. Structure Description of the Base-Specific RNA Recognition by Msi1 RBD2(109–191)
2.5. Heteronuclear Steady-State {1H}-15N Nuclear Overhauser Effect Measurement
3. Discussion
4. Materials and Methods
4.1. Protein and RNA Preparation
4.2. NMR Spectroscopy
4.3. Structure Calculations
4.4. NMR Titration Experiments
4.5. Heteronuclear Steady-State {1H}-15N Nuclear Overhauser Effect (NOE) Measurements
4.6. Model Structure Generation
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sugiyama-Nakagiri, Y.; Akiyama, M.; Shibata, S.; Okano, H.; Shimizu, H. Expression of RNA-binding protein Musashi in hair follicle development and hair cycle progression. Am. J. Pathol. 2006, 168, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.M.; Fraser, B.A.; Sobinoff, A.P.; Pye, V.J.; Davidson, T.-L.; Siddall, N.A.; Koopman, P.; Hime, G.R.; McLaughlin, E.A. Developmental expression of Musashi-1 and Musashi-2 RNA-binding proteins during spermatogenesis: Analysis of the deleterious effects of dysregulated Expression1. Biol. Reprod. 2014, 90, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, S.-I.; Nakamura, Y.; Yoshida, T.; Shibata, S.; Koike, M.; Takano, H.; Ueda, S.; Uchiyama, Y.; Noda, T.; Okano, H. RNA-binding protein Musashi family: Roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. USA 2002, 99, 15194–15199. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Tokunaga, A.; Yoshida, T.; Hashimoto, M.; Mikoshiba, K.; Weinmaster, G.; Nakafuku, M.; Okano, H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001, 21, 3888–3900. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Han, C.; Yu, Q.; Guan, Y.; Li, N.; Zhou, J.; Tian, Y.; Zhang, Y. Downregulation of Msi1 suppresses the growth of human colon cancer by targeting p21cip1. Int. J. Oncol. 2015, 46, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Cui, J.; Quan, M.; Xie, D.; Jia, Z.; Wei, D.; Wang, L.; Gao, Y.; Ma, Q.; Xie, K. The novel KLF4/MSI2 signaling pathway regulates growth and metastasis of pancreatic cancer. Clin. Cancer Res. 2017, 23, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Kanemura, Y.; Yamasaki, M.; Kanemura, Y.; Mori, K.; Fujikawa, H.; Hayashi, H.; Nakano, A.; Matsumoto, T.; Tamura, K.; Arita, N.; et al. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation 2001, 68, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-H.; Jeong, K.J.; Kim, W.Y.; Lee, H.J.; Gong, G.; Suh, N.; Győrffy, B.; Kim, S.; Jeong, S.-Y.; Mills, G.B.; et al. Musashi RNA-binding protein 2 regulates estrogen receptor 1 function in breast cancer. Oncogene 2017, 36, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Kudinov, A.E.; Deneka, A.; Nikonova, A.S.; Beck, T.N.; Ahn, Y.-H.; Liu, X.; Martinez, C.F.; Schultz, F.A.; Reynolds, S.; Yang, D.-H.; et al. Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 6955–6960. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; An, S.; Choi, Y.M.; Lee, J.; Ahn, K.J.; Lee, J.H.; Kim, T.J.; An, I.-S.; Bae, S. Musashi-2 is a novel regulator of paclitaxel sensitivity in ovarian cancer cells. Int. J. Oncol. 2016, 49, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Penalva, L.O.; Yuan, H.; Linnoila, R.I.; Lu, J.; Okano, H.; Glazer, R.I. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol. Cancer 2010, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, W.; Wang, L.; Kazobinka, G.; Han, X.; Li, B.; Hou, T. Musashi-2 promotes migration and invasion in bladder cancer via activation of the JAK2/STAT3 pathway. Lab. Investig. 2016, 96, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Kharas, M.G.; Lengner, C.J.; Al-Shahrour, F.; Bullinger, L.; Ball, B.; Zaidi, S.; Morgan, K.; Tam, W.; Paktinat, M.; Okabe, R.; et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010, 16, 903–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Kwon, H.Y.; Zimdahl, B.; Congdon, K.L.; Blum, J.; Lento, W.E.; Zhao, C.; Lagoo, A.; Gerrard, G.; Foroni, L.; et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010, 466, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Griner, L.N.; Reuther, G.W. Aggressive myeloid leukemia formation is directed by the musashi 2/numb pathway. Cancer Biol. Ther. 2010, 10, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ye, A.; Zhang, Y.; Cai, Z.; Wang, W.; Sun, L.; Jiang, S.; Wu, J.; Yu, K.; Zhang, S. Musashi-2 silencing exerts potent activity against acute myeloid leukemia and enhances chemosensitivity to daunorubicin. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.Y.; Bajaj, J.; Ito, T.; Blevins, A.; Konuma, T.; Weeks, J.; Lytle, N.K.; Koechlein, C.S.; Rizzieri, D.; Chuah, C.; et al. Tetraspanin 3 is required for the development and propagation of acute myelogenous leukemia. Cell Stem Cell 2015, 17, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Nagata, T.; Tsuda, K.; Kobayashi, N.; Imai, T.; Okano, H.; Yamazaki, T.; Katahira, M. Structure of Musashi1 in a complex with target RNA: The role of aromatic stacking interactions. Nucleic Acids Res. 2012, 40, 3218–3231. [Google Scholar] [CrossRef] [PubMed]
- Zearfoss, N.R.; Deveau, L.M.; Clingman, C.C.; Schmidt, E.; Johnson, E.S.; Massi, F.; Ryder, S.P. A conserved three-nucleotide core motif defines musashi RNA binding specificity. J. Biol. Chem. 2014, 289, 35530–35541. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Imai, T.; Imataka, H.; Tsujimoto, M.; Matsumoto, K.; Okano, H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008, 181, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Okada, Y.; Imai, T.; Iwanami, A.; Mischel, P.S.; Okano, H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 2011, 286, 16121–16130. [Google Scholar] [CrossRef] [PubMed]
- Cragle, C.; MacNicol, A.M. Musashi protein-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germ line development defective-2. J. Biol. Chem. 2014, 289, 14239–14251. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, S.; Nakamura, Y.; Satoh, H.; Okano, H. Rna-binding protein Musashi2: Developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J. Neurosci. 2001, 21, 8091–8107. [Google Scholar] [PubMed]
- Battelli, C.; Nikopoulos, G.N.; Mitchell, J.G.; Verdi, J.M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell. Neurosci. 2006, 31, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Vo, D.T.; Subramaniam, D.; Remke, M.; Burton, T.L.; Uren, P.J.; Gelfond, J.A.; de Sousa Abreu, R.; Burns, S.C.; Qiao, M.; Suresh, U.; et al. The RNA-binding protein musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am. J. Pathol. 2012, 181, 1762–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, Y.; Li, F.; Lambert, N.J.; Sokol, E.S.; Tam, W.L.; Cheng, A.W.; Airoldi, E.M.; Lengner, C.J.; Gupta, P.B.; Yu, Z.; et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. Elife 2014, 3, e03915. [Google Scholar] [CrossRef] [PubMed]
- Afroz, T.; Cienikova, Z.; Cléry, A.; Allain, F.H.T. Chapter Nine-one, two, three, four! How multiple RRMs read the genome sequence. Methods Enzymol. 2015, 558, 235–278. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Kanno, R.; Kurihara, Y.; Uesugi, S.; Imai, T.; Sakakibara, S.; Okano, H.; Katahira, M. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J. Mol. Biol. 1999, 287, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Miyanoiri, Y.; Kobayashi, H.; Imai, T.; Watanabe, M.; Nagata, T.; Uesugi, S.; Okano, H.; Katahira, M. Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, Musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics. J. Biol. Chem. 2003, 278, 41309–41315. [Google Scholar] [CrossRef] [PubMed]
- Iwaoka, R.; Nagata, T.; Tsuda, K.; Imai, T.; Okano, H.; Kobayashi, N.; Katahira, M. Backbone and side chain assignments of the second RNA-binding domain of Musashi-1 in its free form and in complex with 5-mer RNA. status (manuscript in preparation).
- Kessler, M.M.; Henry, M.F.; Shen, E.; Zhao, J.; Gross, S.; Silver, P.A.; Moore, C.L. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3’-end formation in yeast. Genes Dev. 1997, 11, 2545–2556. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cañadillas, J.M. Grabbing the message: Structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006, 25, 3167–3178. [Google Scholar] [CrossRef] [PubMed]
- Spears, E.; Neufeld, K.L. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J. Biol. Chem. 2011, 286, 4946–4950. [Google Scholar] [CrossRef] [PubMed]
- Horisawa, K.; Imai, T.; Okano, H.; Yanagawa, H. 3′-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 2009, 583, 2429–2434. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-M.; Deering, R.P.; Lu, Y.; Tivnan, P.; Lianoglou, S.; Al-Shahrour, F.; Ebert, B.L.; Hacohen, N.; Leslie, C.; Daley, G.Q.; et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J. Exp. Med. 2014, 211, 71–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Vandivier, L.; Parada, K.; Woo, D.H.; Wang, S.; Naqvi, A.S.; Rao, S.; et al. The Msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Rep. 2015, 13, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Parada, K.; Rao, S.; Minuesa, G.; Katz, Y.; Gregory, B.D.; et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat. Commun. 2015, 6, 6517. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Lin, L.-T.; Wang, M.-L.; Lee, S.-H.; Tsai, M.-L.; Tsai, C.-C.; Liu, W.-H.; Chen, T.-C.; Yang, Y.-P.; Lee, Y.-Y.; et al. Musashi-1 regulates AKT-derived IL-6 autocrinal/paracrinal malignancy and chemoresistance in glioblastoma. Oncotarget 2016, 7, 42485–42501. [Google Scholar] [CrossRef] [PubMed]
- Chiou, G.-Y.; Yang, T.-W.; Huang, C.-C.; Tang, C.-Y.; Yen, J.-Y.; Tsai, M.-C.; Chen, H.-Y.; Fadhilah, N.; Lin, C.-C.; Jong, Y.-J. Musashi-1 promotes a cancer stem cell lineage and chemoresistance in colorectal cancer cells. Sci. Rep. 2017, 7, 2172. [Google Scholar] [CrossRef] [PubMed]
- Muto, J.; Imai, T.; Ogawa, D.; Nishimoto, Y.; Okada, Y.; Mabuchi, Y.; Kawase, T.; Iwanami, A.; Mischel, P.S.; Saya, H.; et al. RNA-binding protein musashi1 modulates glioma cell growth through the post-transcriptional regulation of notch and PI 3 kinase/Akt signaling pathways. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Choi, Y.M.; Kim, K.B.; Lee, J.H.; Chun, Y.K.; An, I.S.; An, S.; Bae, S. DBC2/RhoBTB2 functions as a tumor suppressor protein via Musashi-2 ubiquitination in breast cancer. Oncogene 2017, 36, 2802–2912. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.G.; Lytle, N.K.; Jaquish, D.V.; Park, F.D.; Ito, T.; Bajaj, J.; Koechlein, C.S.; Zimdahl, B.; Yano, M.; Kopp, J.L.; et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature 2016, 534, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Appelman, C.; Smith, A.R.; Yu, J.; Larsen, S.; Marquez, R.T.; Liu, H.; Wu, X.; Gao, P.; Roy, A.; et al. Natural product (−)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol. Oncol. 2015, 9, 1406–1420. [Google Scholar] [CrossRef] [PubMed]
- Clingman, C.C.; Deveau, L.M.; Hay, S.A.; Genga, R.M.; Shandilya, S.M.D.; Massi, F.; Ryder, S.P. Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. Elife 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Iwahara, J.; Koshiba, S.; Tomizawa, T.; Tochio, N.; Güntert, P.; Kigawa, T.; Yokoyama, S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR 2007, 39, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Harano, Y.; Tochio, N.; Nakatani, E.; Kigawa, T.; Yokoyama, S.; Mading, S.; Ulrich, E.L.; Markley, J.L.; Akutsu, H.; et al. An automated system designed for large scale NMR data deposition and annotation: Application to over 600 assigned chemical shift data entries to the BioMagResBank from the Riken Structural Genomics/Proteomics Initiative internal database. J. Biomol. NMR 2012, 53, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Blevins, R.A. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Kneller, D.G. Sparky—NMR Assignment and Integration Software. Available online: https://www.cgl.ucsf.edu/home/sparky/ (accessed on 24 June 2017).
- Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef] [PubMed]
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program Dyana. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, T.; Güntert, P.; Wüthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002, 319, 209–227. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.; Garrett, D.S.; March, C.J.; Frieden, E.A.; Gronenborn, A.M.; Clore, G.M. The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry 1993, 32, 6744–6762. [Google Scholar] [CrossRef] [PubMed]
- Amber Home Page. Available online: http://ambermd.org (accessed on 24 June 2017).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Farrow, N.A.; Muhandiram, R.; Singer, A.U.; Pascal, S.M.; Kay, C.M.; Gish, G.; Shoelson, S.E.; Pawson, T.; Forman-Kay, J.D.; Kay, L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N-NMR relaxation. Biochemistry 1994, 33, 5984–6003. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |







| Free Msi1(109–200)(109–200) | Msi1(109–200):r(GUAGU) Complex | |
|---|---|---|
| NMR Restraints | ||
| Number of NOE distance restraints | 1116 | 1351 |
| Intraresidue | 315 | 341 |
| Sequential (|i − j| = 1) | 300 | 324 |
| Medium-range (1 < |i − j| < 5) | 165 | 191 |
| Long-range (|i − j| ≥ 5) | 336 | 468 |
| Proteins—RNA Intermolecular | NA | 26 |
| RNA intramolecule | NA | 1 |
| Hydrogen bond restraints a | 27 | 27 |
| Dihedral angle restraints | ||
| φ angle | 32 | 0 |
| χ1 and χ2 angles | 19 | 19 |
| Structure statistics (20 structures) | ||
| AMBER energies (kcal/mol) | ||
| Mean AMBER energy | −2780 | −3800 |
| Mean restraints violation energy | 4.698 | 4.359 |
| Ramachandran plot statistics (%) | ||
| Residues in most favored regions | 91.5 | 91.5 |
| Residues in additionally allowed regions | 7.0 | 7.0 |
| Residues in generously allowed regions | 1.3 | 0.2 |
| sidues in disallowed regions | 0.2 | 1.4 |
| Average RMSD to mean structure (Å) | ||
| Protein backbone | 0.55 ± 0.11 b | 0.74 ± 0.28 c (0.35 ± 0.07 b) |
| Protein heavy atoms | 1.45 ± 0.21 b | 1.45 ± 0.29 c (1.13 ± 0.15 b) |
| Protein heavy atoms and r(UAG) | NA | 1.42 ± 0.28 c (1.13 ± 0.15 b) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwaoka, R.; Nagata, T.; Tsuda, K.; Imai, T.; Okano, H.; Kobayashi, N.; Katahira, M. Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA. Molecules 2017, 22, 1207. https://doi.org/10.3390/molecules22071207
Iwaoka R, Nagata T, Tsuda K, Imai T, Okano H, Kobayashi N, Katahira M. Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA. Molecules. 2017; 22(7):1207. https://doi.org/10.3390/molecules22071207
Chicago/Turabian StyleIwaoka, Ryo, Takashi Nagata, Kengo Tsuda, Takao Imai, Hideyuki Okano, Naohiro Kobayashi, and Masato Katahira. 2017. "Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA" Molecules 22, no. 7: 1207. https://doi.org/10.3390/molecules22071207
APA StyleIwaoka, R., Nagata, T., Tsuda, K., Imai, T., Okano, H., Kobayashi, N., & Katahira, M. (2017). Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA. Molecules, 22(7), 1207. https://doi.org/10.3390/molecules22071207