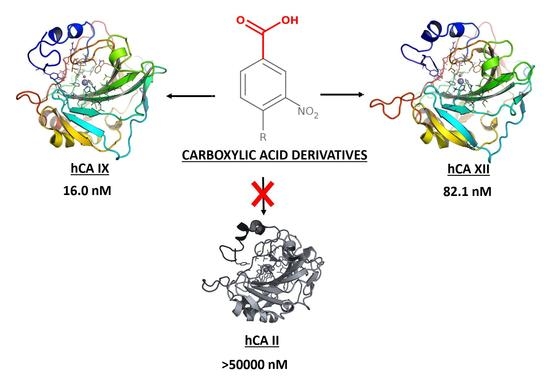
Potent and Selective Carboxylic Acid Inhibitors of Tumor-Associated Carbonic Anhydrases IX and XII
Abstract
:1. Introduction
2. Results and Discussion
2.1. Substructure Search and Ligands Selection
2.2. Carbonic Anhydrase Inhibition
- (i)
- All tested compound are poorly active against cytosolic hCA I and hCA II, as Ki values were significantly higher with respect to the reference compounds AAZ and GV2-20. The drop of inhibitory potency compared to GV2-20 is particularly marked against hCA II, where the most potent inhibitor of the series, namely compound 1, has a Ki around 54-fold higher than that of AAZ and around 10-fold higher than that of GV2-20. Sub-micromolar inhibition of hCA I was observed only for compounds 1 and 8, bearing respectively a 1,3,4-oxadiazole and a 1,3,4-thiadiazole moiety in the tail, while 1 was the only hCA I inhibitor with sub-micromolar Ki. Modifications to the heterocycle such as the tetrazole in 17 proved ineffective for hCA II inhibition. Other tested compounds showed Ki in the micromolar range, or proved inactive at all against these cytosolic hCA isoforms.
- (ii)
- Similar to the reference inhibitor GV2-20 but different from AAZ, tested compounds proved generally inactive also against hCA IV.
- (iii)
- Compared to GV2-20, most of tested compounds showed a significant improvement of hCA VA inhibition. Notably, the most potent hCA VA inhibitor of the series was 19, in which the carboxylic acid is replaced by the nitrile function thus opening new venues for the design of hCA VA-specific inhibitors. Other compounds with a low Ki against this isoform are 10, 12, 13, 15, and 16 that also show a certain degree of specificity for hCA VA particularly compared to cytosolic hCA I and hCA II, hCA IV, and hCA VII.
- (iv)
- Most of tested compound were poorly active against hCA VII that, in contrast, was efficiently inhibited by GV2-20 and AAZ. As already observed for hCA I and hCA II, compound 1 was the most potent inhibitor of hCA VII, even though the chemically-related compound 8 proved inactive. Compared to GV2-20, compound 1 showed around 37-fold drop of inhibitory potency against hCA VII. Moreover, 5, 13, and 19 that are structurally-related to GV2-20 proved inactive. This suggests that subtle modifications to GV2-20 scaffold have a dramatic impact on hCA VII inhibition, and that the 3,5-dinitrobenzoic acid is required to target this hCA isoform.
- (v)
- Tumor-associated isoforms hCA IX and hCA XII were potently inhibited by most of tested compounds. It is worth noting that some compounds showed stronger or at least similar inhibition of hCA IX compared to the reference compounds AAZ and GV2-20, see for example 6–8, 10, 14–17, whereas none of them was able to overtake AAZ or GV2-20 against hCA XII. The most potent inhibitor of hCA IX was 10, which also inhibited hCA XII in the low nanomolar range. Unique among other is the behavior of compounds 6 and 7, which are low nanomolar inhibitors of CA IX with potency comparable to AAZ, although they are not active against hCA XII. Of note, 7 is the lowest MW compound of the test-set, thus becoming the selective hCA IX inhibitor endowed with the highest ligand efficiency identified in this work [55]. Therefore, its structure could be easily expanded by rational design with the aim to optimize physicochemical and pharmacological properties up to the level of confirmed lead or preclinical candidate. Finally, since hCA II is the most physiologically abundant hCA isoform, and is generally referred as the major causes of CAIs side-effects [1], the hCA II/hCA IX and hCA II/hCA XII selectivity indexes are showed in Table 1. Notably, the most potent inhibitors of tumor-associated hCA IX and hCA XII isoforms are also significantly selective with respect to the cytosolic hCA II. While all compounds showed a greater selectivity than the reference inhibitors GV2-20 and AAZ, compound 1 showed the weakest specificity for tumor-associated hCAs, whereas 10 emerged as the most selective one.
- (vi)
- The GV2-20 derivative 18 that is deprived of the nitro group in the head portion is poorly active in most hCA isoforms, with the only exception of hCA IX, for which it showed a Ki of 72.8 nM. Based on the comparison with the hCAs inhibition profile of GV2-20, we may speculate that at least one nitro group in the head portion is essential for the efficient inhibition of most hCAs. Compound 19 that bears a nitrile instead of carboxylic acid group, shares a similar hCAs inhibition profile as 18 with the exception of a stronger inhibition of hCA VA (Ki = 92.5 nM).
2.3. Molecular Modeling Study
3. Experimental Protocols
3.1. Selection of GV2-20 Derivatives and Molecular Modeling
3.2. CA Inhibition
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lomelino, C.L.; Supuran, C.T.; McKenna, R. Non-Classical Inhibition of Carbonic Anhydrase. Int. J. Mol. Sci. 2016, 17, 1150. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases—An overview. Curr. Pharm. Des. 2008, 14, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Masini, E.; Carta, F.; Scozzafava, A.; Supuran, C.T. Antiglaucoma carbonic anhydrase inhibitors: A patent review. Expert Opin. Ther. Pat. 2013, 23, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Kondeti, B.; McKenna, R. Anticonvulsant/antiepileptic carbonic anhydrase inhibitors: A patent review. Expert Opin. Ther. Pat. 2013, 23, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Buzas, G.M.; Supuran, C.T. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puscas (1932–2015). J. Enzyme Inhib. Med. Chem. 2016, 31, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Sein, K.K.; Aikawa, M. The pivotal role of carbonic anhydrase in malaria infection. Med. Hypotheses 1998, 50, 19–23. [Google Scholar] [CrossRef]
- Saad, A.E.; Ashour, D.S.; Abou Rayia, D.M.; Bedeer, A.E. Carbonic anhydrase enzyme as a potential therapeutic target for experimental trichinellosis. Parasitol. Res. 2016, 115, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Carradori, S.; Mollica, A.; De Monte, C.; Ganese, A.; Supuran, C.T. Nitric oxide donors and selective carbonic anhydrase inhibitors: A dual pharmacological approach for the treatment of glaucoma, cancer and osteoporosis. Molecules 2015, 20, 5667–5679. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Di Fiore, A.; De Simone, G. Carbonic anhydrase inhibitors as emerging drugs for the treatment of obesity. Expert Opin. Emerg. Drugs 2008, 13, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic Anhydrase Inhibition and the Management of Hypoxic Tumors. Metabolites 2017, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sharma, V.; Bua, S.; Supuran, C.T.; Sharma, P.K. Synthesis and biological evaluation of benzenesulphonamide-bearing 1,4,5-trisubstituted-1,2,3-triazoles possessing human carbonic anhydrase I, II, IV, and IX inhibitory activity. J. Enzyme Inhib. Med. Chem. 2017, 32, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Durante, M.; Di Leva, F.S.; Cosconati, S.; Masini, E.; Scozzafava, A.; Novellino, E.; Supuran, C.T.; Carta, F. Monothiocarbamates Strongly Inhibit Carbonic Anhydrases in Vitro and Possess Intraocular Pressure Lowering Activity in an Animal Model of Glaucoma. J. Med. Chem. 2016, 59, 5857–5867. [Google Scholar] [CrossRef] [PubMed]
- Scozzafava, A.; Supuran, C.T. Glaucoma and the applications of carbonic anhydrase inhibitors. Subcell Biochem. 2014, 75, 349–359. [Google Scholar] [PubMed]
- Nocentini, A.; Ceruso, M.; Carta, F.; Supuran, C.T. 7-Aryl-triazolyl-substituted sulfocoumarins are potent, selective inhibitors of the tumor-associated carbonic anhydrase IX and XII. J. Enzyme Inhib. Med. Chem. 2016, 31, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Karali, N.; Akdemir, A.; Goktas, F.; Eraslan Elma, P.; Angeli, A.; Kizilirmak, M.; Supuran, C.T. Novel sulfonamide-containing 2-indolinones that selectively inhibit tumor-associated alpha carbonic anhydrases. Bioorg. Med. Chem. 2017, 25, 3714–3718. [Google Scholar] [CrossRef] [PubMed]
- D’Ascenzio, M.; Guglielmi, P.; Carradori, S.; Secci, D.; Florio, R.; Mollica, A.; Ceruso, M.; Akdemir, A.; Sobolev, A.P.; Supuran, C.T. Open saccharin-based secondary sulfonamides as potent and selective inhibitors of cancer-related carbonic anhydrase IX and XII isoforms. J. Enzyme Inhib. Med. Chem. 2017, 32, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Scozzafava, A.; Supuran, C.T.; Carta, F. Antiobesity carbonic anhydrase inhibitors: A literature and patent review. Expert Opin. Ther. Pat. 2013, 23, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Queen, A.; Khan, P.; Azam, A.; Hassan, M.I. Understanding the role and mechanism of carbonic anhydrase V in obesity and its therapeutic implications. Curr. Protein Pept. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibitors as emerging drugs for the treatment of obesity. Expert Opin. Emerg. Drugs 2012, 17, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Supuran, C.T. QSARs on human carbonic anhydrase VA and VB inhibitors of some new not yet synthesized, substituted aromatic/heterocyclic sulphonamides as anti-obesity agent. J. Enzyme Inhib. Med. Chem. 2012, 27, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Ruusuvuori, E.; Huebner, A.K.; Kirilkin, I.; Yukin, A.Y.; Blaesse, P.; Helmy, M.; Kang, H.J.; El Muayed, M.; Hennings, J.C.; Voipio, J.; et al. Neuronal carbonic anhydrase VII provides GABAergic excitatory drive to exacerbate febrile seizures. EMBO J. 2013, 32, 2275–2286. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibition and the management of neuropathic pain. Expert Rev. Neurother. 2016, 16, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Carta, F.; Di Cesare Mannelli, L.; Pinard, M.; Ghelardini, C.; Scozzafava, A.; McKenna, R.; Supuran, C.T. A class of sulfonamide carbonic anhydrase inhibitors with neuropathic pain modulating effects. Bioorg. Med. Chem. 2015, 23, 1828–1840. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016, 473, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Cau, Y.; Vignaroli, G.; Laurenzana, I.; Caivano, A.; Vullo, D.; Supuran, C.T.; Botta, M. Hit Recycling: Discovery of a Potent Carbonic Anhydrase Inhibitor by in Silico Target Fishing. ACS Chem. Biol. 2015, 10, 1964–1999. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Vignaroli, G.; Cau, Y.; Dinic, J.; Hill, R.; Rossi, M.; Colecchia, D.; Pesic, M.; Link, W.; Chiariello, M.; et al. Discovery of 14-3-3 protein-protein interaction inhibitors that sensitize multidrug-resistant cancer cells to doxorubicin and the Akt inhibitor GSK690693. ChemMedChem 2014, 9, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Cau, Y.; Valensin, D.; Mori, M.; Draghi, S.; Botta, M. Structure, function, involvement in diseases and targeting of 14-3-3 proteins: An update. Curr. Med. Chem. 2017, 25, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Vignaroli, G.; Botta, M. Small molecules modulation of 14-3-3 protein-protein interactions. Drug Discov. Today Technol. 2013, 10, e541–e547. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. How many carbonic anhydrase inhibition mechanisms exist? J. Enzyme Inhib. Med. Chem. 2016, 31, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Xavier, N.M.; Lucas, S.D.; Jorda, R.; Schwarz, S.; Loesche, A.; Csuk, R.; Oliveira, M.C. Synthesis and Evaluation of the Biological Profile of Novel Analogues of Nucleosides and of Potential Mimetics of Sugar Phosphates and Nucleotides. Synlett 2015, 26, 2663–2672. [Google Scholar] [CrossRef]
- Tullis, J.S.; Laufersweiler, M.J.; VanRens, J.C.; Natchus, M.G.; Bookland, R.G.; Almstead, N.G.; Pikul, S.; De, B.; Hsieh, L.C.; Janusz, M.J.; et al. The development of new carboxylic acid-based MMP inhibitors derived from a cyclohexylglycine scaffold. Bioorg. Med. Chem. Lett. 2001, 11, 1975–1979. [Google Scholar] [CrossRef]
- Fujisawa, T.; Katakura, S.; Odake, S.; Morita, Y.; Yasuda, J.; Yasumatsu, I.; Morikawa, T. Design and synthesis of carboxylate inhibitors for matrix metalloproteinases. Chem. Pharm. Bull. 2001, 49, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.A.; Major Jourden, J.L.; Miller, M.T.; Cohen, S.M. To bind zinc or not to bind zinc: An examination of innovative approaches to improved metalloproteinase inhibition. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 72–94. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Massaro, A.; Calderone, V.; Fragai, M.; Luchinat, C.; Mordini, A. Discovery of a New Class of Potent MMP Inhibitors by Structure-Based Optimization of the Arylsulfonamide Scaffold. ACS Med. Chem. Lett. 2013, 4, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Testa, F.; Gargiulo, A.; Di Iorio, V.; Pierri, R.B.; D’Alterio, F.M.; Corte, M.D.; Surace, E.; Simonelli, F. The role of optical coherence tomography in an atypical case of oculocutaneous albinism: A case report. Case Rep. Ophthalmol. 2012, 3, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Moraca, F.; Deodato, D.; Ferraris, D.M.; Selchow, P.; Sander, P.; Rizzi, M.; Botta, M. Discovery of the first potent and selective Mycobacterium tuberculosis Zmp1 inhibitor. Bioorg. Med. Chem. Lett. 2014, 24, 2508–2511. [Google Scholar] [CrossRef] [PubMed]
- Cirigliano, A.; Menta, S.; Mori, M.; Licursi, V.; Danovska, S.; Vapore, V.; Serino, G.; Pick, E.; Botta, B.; Negri, R.; et al. S. cerevisiae as a tool to select inhibitors of the deneddylating activity of the COP9 signalosome. J. Enzyme Inhib. Med. Chem. 2016, 31, 1632–1637. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.A.; Fulla bgar, J.L.; Miller, M.T.; Cohen, S.M. Identifying chelators for metalloprotein inhibitors using a fragment-based approach. J. Med. Chem. 2011, 54, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin. Drug. Discov. 2017, 12, 61–88. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, P.A.; Kumaran, D.; Swaminathan, S.; Das, A.K. A novel acetate-bound complex of human carbonic anhydrase II. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 163–166. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, K.; Carradori, S.; Monti, S.M.; Buonanno, M.; Secci, D.; Vullo, D.; Supuran, C.T.; De Simone, G. Out of the active site binding pocket for carbonic anhydrase inhibitors. Chem. Commun. 2015, 51, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Cohen, S.M. Nucleophile recognition as an alternative inhibition mode for benzoic acid based carbonic anhydrase inhibitors. Chem. Commun. 2012, 48, 5259–5261. [Google Scholar] [CrossRef] [PubMed]
- Gueneau, E.; Dherin, C.; Legrand, P.; Tellier-Lebegue, C.; Gilquin, B.; Bonnesoeur, P.; Londino, F.; Quemener, C.; Le Du, M.H.; Marquez, J.A.; et al. Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat. Struct. Mol. Biol. 2013, 20, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Alterio, V.; Langella, E.; Viparelli, F.; Vullo, D.; Ascione, G.; Dathan, N.A.; Morel, F.M.; Supuran, C.T.; De Simone, G.; Monti, S.M. Structural and inhibition insights into carbonic anhydrase CDCA1 from the marine diatom Thalassiosira weissflogii. Biochimie 2012, 94, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Cau, Y.; Mori, M.; Supuran, C.T.; Botta, M. Mycobacterial carbonic anhydrase inhibition with phenolic acids and esters: Kinetic and computational investigations. Org. Biomol. Chem. 2016, 14, 8322–8330. [Google Scholar] [CrossRef] [PubMed]
- Maresca, A.; Temperini, C.; Pochet, L.; Masereel, B.; Scozzafava, A.; Supuran, C.T. Deciphering the mechanism of carbonic anhydrase inhibition with coumarins and thiocoumarins. J. Med. Chem. 2010, 53, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Maresca, A.; Temperini, C.; Pochet, L.; Masereel, B.; Scozzafava, A.; Supuran, C.T. Correction to Deciphering the Mechanism of Carbonic Anhydrase Inhibition with Coumarins and Thiocoumarins. J. Med. Chem. 2015, 58, 5689. [Google Scholar] [CrossRef] [PubMed]
- Berrino, E.; Bua, S.; Mori, M.; Botta, M.; Murthy, V.S.; Vijayakumar, V.; Tamboli, Y.; Bartolucci, G.; Mugelli, A.; Cerbai, E.; et al. Novel Sulfamide-Containing Compounds as Selective Carbonic Anhydrase I Inhibitors. Molecules 2017, 22, 1049. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Structure-based drug discovery of carbonic anhydrase inhibitors. J. Enzyme Inhib. Med. Chem. 2012, 27, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Carta, F.; Supuran, C.T.; Scozzafava, A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med. Chem. 2014, 6, 1149–1165. [Google Scholar] [CrossRef] [PubMed]
- Rouffet, M.; Cohen, S.M. Emerging trends in metalloprotein inhibition. Dalton Trans. 2011, 40, 3445–3454. [Google Scholar] [CrossRef] [PubMed]
- Luisi, G.; Angelini, G.; Gasbarri, C.; Laghezza, A.; Agamennone, M.; Loiodice, F.; Supuran, C.T.; Campestre, C.; Tortorella, P. Dual targeting of cancer-related human matrix metalloproteinases and carbonic anhydrases by chiral N-(biarylsulfonyl)-phosphonic acids. J. Enzyme Inhib. Med. Chem. 2017, 32, 1260–1264. [Google Scholar] [CrossRef] [PubMed]
- Khalifah, R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971, 246, 2561–2573. [Google Scholar] [PubMed]
- Hopkins, A.L.; Keseru, G.M.; Leeson, P.D.; Rees, D.C.; Reynolds, C.H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 2014, 13, 105–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. OMEGA 2.5.1.4; OpenEye Scientific Software: Santa Fe, NM, USA; Available online: http://www.eyesopen.com (accessed on 27 October 2017).
- Hawkins, P.C.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Mod. 2010, 50, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Alterio, V.; Hilvo, M.; Di Fiore, A.; Supuran, C.T.; Pan, P.; Parkkila, S.; Scaloni, A.; Pastorek, J.; Pastorekova, S.; Pedone, C.; et al. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc. Natl. Acad. Sci. USA 2009, 106, 16233–16238. [Google Scholar] [CrossRef] [PubMed]
- Whittington, D.A.; Waheed, A.; Ulmasov, B.; Shah, G.N.; Grubb, J.H.; Sly, W.S.; Christianson, D.W. Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc. Natl. Acad. Sci. USA 2001, 98, 9545–9550. [Google Scholar] [CrossRef] [PubMed]
- QUACPAC 1.7.0.2; OpenEye Scientific Software: Santa Fe, NM, USA; Available online: http://www.eyesopen.com (accessed on 27 October 2017).
- Case, D.A.; CeruttiD, D.S.; Cheatham, T.E.; Darden, T.A., III; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER 2017; University of California: San Francisco, CA, USA, 2017. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.P. Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method. Proteins 2001, 45, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; Version 2.0; Schrödinger, LLC: New York, NY, USA, 2017. [Google Scholar]
- Schrödinger Release 2017-3: Maestro; Schrödinger, LLC: New York, NY, USA, 2017.
- Scozzafava, A.; Briganti, F.; Mincione, G.; Menabuoni, L.; Mincione, F.; Supuran, C.T. Carbonic anhydrase inhibitors: Synthesis of water-soluble, aminoacyl/dipeptidyl sulfonamides possessing long-lasting intraocular pressure-lowering properties via the topical route. J. Med. Chem. 1999, 42, 3690–3700. [Google Scholar] [CrossRef] [PubMed]
- Puccetti, L.; Fasolis, G.; Cecchi, A.; Winum, J.Y.; Gamberi, A.; Montero, J.L.; Scozzafava, A.; Supuran, C.T. Carbonic anhydrase inhibitors: Synthesis and inhibition of cytosolic/tumor-associated carbonic anhydrase isozymes I, II, and IX with sulfonamides incorporating thioureido-sulfanilyl scaffolds. Bioorg. Med. Chem. Lett. 2005, 15, 2359–2364. [Google Scholar] [CrossRef] [PubMed]
- Scozzafava, A.; Menabuoni, L.; Mincione, F.; Supuran, C.T. Carbonic anhydrase inhibitors. A general approach for the preparation of water-soluble sulfonamides incorporating polyamino-polycarboxylate tails and of their metal complexes possessing long-lasting, topical intraocular pressure-lowering properties. J. Med. Chem. 2002, 45, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Nicolae, A.; Popescu, A. Carbonic anhydrase inhibitors. Part 35. Synthesis of Schiff bases derived from sulfanilamide and aromatic aldehydes: The first inhibitors with equally high affinity towards cytosolic and membrane-bound isozymes. Eur. J. Med. Chem. 1996, 31, 431–438. [Google Scholar] [CrossRef]
- Sarikaya, S.B.O.; Topal, F.; Senturk, M.; Gulcin, I.; Supuran, C.T. In vitro inhibition of alpha-carbonic anhydrase isozymes by some phenolic compounds. Bioorg. Med. Chem. Lett. 2011, 21, 4259–4262. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |





| Cmpd | Ki (nM) a | Selectivity Index b | |||||||
|---|---|---|---|---|---|---|---|---|---|
| hCA I | hCA II | hCA IV | hCA VA | hCA VII | hCA IX | hCA XII | hCA II/hCA IX | hCA II/hCA XII | |
| 1 | 700 | 655 | 5090 | 826 | 325 | 55.1 | 27.3 | 11.88 | 23.99 |
| 2 | 5645 | 7240 | >50,000 | 1352 | >50,000 | 106 | >50,000 | 68.3 | - |
| 3 | >50,000 | 7300 | >50,000 | 1314 | 840 | 209 | >50,000 | 34.9 | - |
| 4 | 6020 | 2630 | >50,000 | >50,000 | >50,000 | 172 | >50,000 | 15.3 | - |
| 5 | 8150 | >50,000 | >50,000 | 1163 | >50,000 | 153 | 863 | >326.8 | >57.9 |
| 6 | 8210 | 8050 | >50,000 | >50,000 | >50,000 | 30.6 | >50,000 | 263.1 | - |
| 7 | >50,000 | 7310 | >50,000 | 1456 | >50,000 | 27.6 | >50,000 | 264.9 | - |
| 8 | 6450 | 801 | 8800 | >50,000 | >50,000 | 26.3 | 912 | 30.4 | 0.9 |
| 9 | >50,000 | 8500 | 8270 | >50,000 | 762 | 165 | 753 | 51.5 | 11.3 |
| 10 | 6650 | >50,000 | >50,000 | 142 | 665 | 16.0 | 82.1 | >3125 | >609.01 |
| 11 | >50,000 | >50,000 | >50,000 | 356 | >50,000 | 58.3 | 778 | >857.6 | >64.3 |
| 12 | >50,000 | >50,000 | >50,000 | 144 | >50,000 | 82.0 | 91.9 | >609.0 | >544.1 |
| 13 | >50,000 | >50,000 | >50,000 | 117 | >50,000 | 106 | 633 | >471.7 | >79.0 |
| 14 | >50,000 | >50,000 | >50,000 | 449 | >50,000 | 23.0 | 482 | >2174.0 | >103.7 |
| 15 | >50,000 | >50,000 | 9200 | 110 | >50,000 | 24.7 | 724 | >2024.3 | >69.1 |
| 16 | >50,000 | >50,000 | 6030 | 154 | 1733 | 30.1 | 352 | >1661.1 | >142.0 |
| 17 | >50,000 | >50,000 | 3560 | 739 | >50,000 | 23.4 | 441 | >2136.8 | >113.4 |
| 18 | 2730 | 7630 | >50,000 | 262 | >50,000 | 72.8 | 629 | 104.8 | 12.1 |
| 19 | >50,000 | >50,000 | >50,000 | 92.5 | >50,000 | 79.1 | 648 | >632.1 | 77.2 |
| GV2-20 | 352 | 67.3 | 7660 | 895 | 8.7 | 42.3 | 9.6 | 1.59 | 7.01 |
| AAZ | 250 | 12.1 | 74.3 | 63.5 | 2.6 | 25.0 | 5.7 | 0.48 | 2.12 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cau, Y.; Vullo, D.; Mori, M.; Dreassi, E.; Supuran, C.T.; Botta, M. Potent and Selective Carboxylic Acid Inhibitors of Tumor-Associated Carbonic Anhydrases IX and XII. Molecules 2018, 23, 17. https://doi.org/10.3390/molecules23010017
Cau Y, Vullo D, Mori M, Dreassi E, Supuran CT, Botta M. Potent and Selective Carboxylic Acid Inhibitors of Tumor-Associated Carbonic Anhydrases IX and XII. Molecules. 2018; 23(1):17. https://doi.org/10.3390/molecules23010017
Chicago/Turabian StyleCau, Ylenia, Daniela Vullo, Mattia Mori, Elena Dreassi, Claudiu T. Supuran, and Maurizio Botta. 2018. "Potent and Selective Carboxylic Acid Inhibitors of Tumor-Associated Carbonic Anhydrases IX and XII" Molecules 23, no. 1: 17. https://doi.org/10.3390/molecules23010017
APA StyleCau, Y., Vullo, D., Mori, M., Dreassi, E., Supuran, C. T., & Botta, M. (2018). Potent and Selective Carboxylic Acid Inhibitors of Tumor-Associated Carbonic Anhydrases IX and XII. Molecules, 23(1), 17. https://doi.org/10.3390/molecules23010017