The Two Isoforms of Lyn Display Different Intramolecular Fuzzy Complexes with the SH3 Domain
Abstract
:1. Introduction
2. Results
2.1. Protein Constructions and NMR Assignments
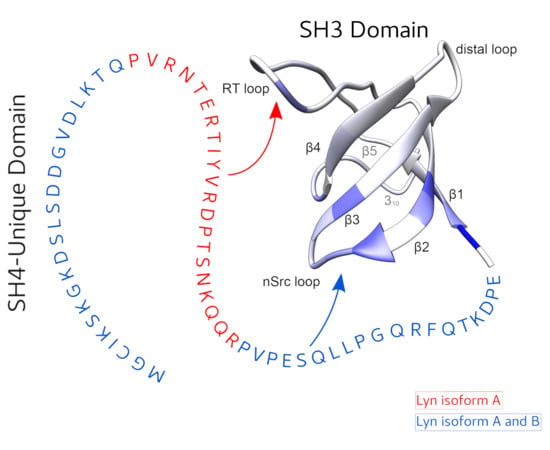
2.2. The Two Isoforms of Lyn form Different Intramolecular Fuzzy Complexes Involving the Unique and SH3 Domains
2.3. The SH4 Domain Also Approaches the SH3 Domain, Although Does Not Show a Direct Interaction
2.4. A Polyproline Peptide Binding to the SH3 Domain Prevents the Specific Interaction of LynA UD with the SH3 Domain
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yeatman, T.J. A renaissance for Src. Nat. Rev. 2004, 4, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Sigal, C.T.; Zhou, W.; Buser, C.A.; McLaughlin, S.; Resh, M.D. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc. Natl. Acad. Sci. USA 1994, 91, 12253–12257. [Google Scholar] [CrossRef] [PubMed]
- Arbesú, M.; Maffei, M.; Cordeiro, T.N.; Teixeira, J.M.C.; Pérez, Y.; Bernadó, P.; Roche, S.; Pons, M. The Unique Domain forms a fuzzy intramolecular complex in Src Family Kinases. Structure 2017, 25, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.W.; Sun, Z.Y.; Blacklow, S.C.; Wagner, G.; Eck, M.J. A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8. Science 2003, 301, 1725–1728. [Google Scholar] [CrossRef] [PubMed]
- Luciano, F.; Ricci, J.-E.; Auberger, P. Cleavage of Fyn and Lyn in their N-terminal unique regions during induction of apoptosis: A new mechanism for Src kinase regulation. Oncogene 2001, 20, 4935–4941. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Mao, S.Y.; Metzger, H. Aggregation of the high-affinity IgE receptor and enhanced activity of p53/56lyn protein-tyrosine kinase. Proc. Natl. Acad. Sci. USA 1994, 91, 11251–11255. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Bolen, J.B.; Ihle, J.N. Hematopoietic cells express two forms of lyn kinase differing by 21 amino acids in the amino terminus. Mol. Cell. Biol. 1991, 11, 2391–2398. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Errico, D.; Yamashita, Y.; Suzuki, R.; Odom, S.; Furumoto, Y.; Yamashita, T.; Rivera, J. Functional analysis of Lyn kinase A and B isoforms reveals redundant and distinct roles in Fc epsilon RI-dependent mast cell activation. J. Immunol. 2010, 184, 5000–5008. [Google Scholar] [CrossRef] [PubMed]
- Niklas, K.J.; Bondos, S.E.; Dunker, K.A.; Newman, S.A. Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Front. Cell Dev. Biol. 2015, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhao, S.; Dunker, A.K. Intrinsically disordered proteins link alternative splicing and post-translational modifications to complex cell signaling and regulation. J. Mol. Biol. 2018, 430, 2342–2359. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, K.; Lebrun, P.; Tompa, P. To be disordered or not to be disordered: Is that still a question for proteins in the cell? Cell. Mol. Life Sci. 2017, 74, 3185–3204. [Google Scholar] [CrossRef] [PubMed]
- Marasco, D.; Scognamiglio, P.L. Identification of inhibitors of biological interactions involving intrinsically disordered proteins. Int. J. Mol. Sci. 2015, 16, 7394–7412. [Google Scholar] [CrossRef] [PubMed]
- Bauer, F.; Schweimer, K.; Meiselbach, H.; Hoffmann, S.; Rösch, P.; Sticht, H. Structural characterization of Lyn-SH3 domain in complex with a herpesviral protein reveals an extended recognition motif that enhances binding affinity. Protein Sci. 2005, 14, 2487–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orekhov, V.Y.; Jaravine, V.A. Analysis of non-uniformly sampled spectra with multi-dimensional decomposition. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka-Kazimierczuk, A.; Koźmiński, W.; Sanderová, H.; Krásný, L. High dimensional and high resolution pulse sequences for backbone resonance assignment of intrinsically disordered proteins. J. Biomol. NMR 2012, 52, 329–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazimierczuk, K.; Zawadzka-Kazimierczuk, A.; Koźmiński, W.A. Non-uniform frequency domain for optimal exploitation of non-uniform sampling. J. Magn. Reson. 2010, 205, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Clore, G.M.; Iwahara, J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009, 109, 4108–4139. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Harrison, S.C.; Eck, M.J. Three-dimensional structure of the tyrosine kinase c-Src. Nature 1997, 385, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Kasahara, C.; Rickles, R.J.; Schreiber, S.L. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc. Natl. Acad. Sci. USA 1995, 92, 12408–12415. [Google Scholar] [CrossRef] [PubMed]
- Moroco, J.A.; Craigo, J.K.; Iacob, R.E.; Wales, T.E.; Engen, J.R.; Smithgall, T.E. Differential sensitivity of Src-family kinases to activation by SH3 domain displacement. PLoS ONE 2014, 9, e105629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbesú, M.; Iruela, G.; Fuentes, H.; Teixeira, J.M.C.; Pons, M. Intramolecular fuzzy interactions involving intrinsically disordered domains. Front. Mol. Biosci. 2018, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Amata, I.; Maffei, M.; Pons, M. Phosphorylation of unique domains of Src family kinases. Front. Genet. 2011, 5, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lock, P.; Ralph, S.; Stanley, E.; Boulet, I.; Ramsay, R.; Dunn, A.R. Two isoforms of murine hck, generated by utilization of alternative translational initiation codons, exhibit different patterns of subcellular localization. Mol. Cell. Biol. 1991, 11, 4363–4370. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMR Pipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Stanek, J.; Koźmiński, W. Iterative algorithm of discrete Fourier transform for processing randomly sampled NMR data sets. J. Biomol. NMR 2010, 47, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Kosiński, K.; Stanek, J.; Górka, M.J.; Żerko, S.; Koźmiński, W. Reconstruction of non-uniformly sampled five-dimensional NMR spectra by signal separation algorithm. J. Biomol. NMR 2017, 68, 129–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SPARKY Version 3.115; Software for Spectra Analyze; University of California: San Francisco, CA, USA, 2000; Available online: https://www.cgl.ucsf.edu/home/sparky (accessed on 30 May 2008).
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.M.C.; Skinner, S.P.; Arbesú, M.; Breeze, A.L.; Pons, M. Farseer-NMR: Automatic treatment, analysis and plotting of large, multi-variable NMR data. J. Biomol. NMR 2018, 71, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ozenne, V.; Bauer, F.; Salmon, L.; Huang, J.R.; Jensen, M.R.; Segard, S.; Bernadó, P.; Charavay, C.; Blackledge, M. Flexible-meccano: A tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 2012, 28, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Plasmids for the expressed constructs are available from the authors. |





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira, J.M.C.; Fuentes, H.; Bielskutė, S.; Gairi, M.; Żerko, S.; Koźmiński, W.; Pons, M. The Two Isoforms of Lyn Display Different Intramolecular Fuzzy Complexes with the SH3 Domain. Molecules 2018, 23, 2731. https://doi.org/10.3390/molecules23112731
Teixeira JMC, Fuentes H, Bielskutė S, Gairi M, Żerko S, Koźmiński W, Pons M. The Two Isoforms of Lyn Display Different Intramolecular Fuzzy Complexes with the SH3 Domain. Molecules. 2018; 23(11):2731. https://doi.org/10.3390/molecules23112731
Chicago/Turabian StyleTeixeira, João M. C., Héctor Fuentes, Stasė Bielskutė, Margarida Gairi, Szymon Żerko, Wiktor Koźmiński, and Miquel Pons. 2018. "The Two Isoforms of Lyn Display Different Intramolecular Fuzzy Complexes with the SH3 Domain" Molecules 23, no. 11: 2731. https://doi.org/10.3390/molecules23112731
APA StyleTeixeira, J. M. C., Fuentes, H., Bielskutė, S., Gairi, M., Żerko, S., Koźmiński, W., & Pons, M. (2018). The Two Isoforms of Lyn Display Different Intramolecular Fuzzy Complexes with the SH3 Domain. Molecules, 23(11), 2731. https://doi.org/10.3390/molecules23112731