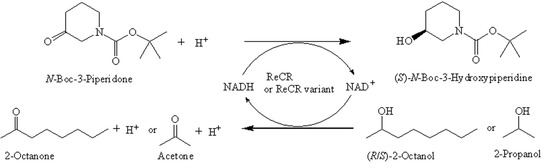
Characterization of a Carbonyl Reductase from Rhodococcus erythropolis WZ010 and Its Variant Y54F for Asymmetric Synthesis of (S)-N-Boc-3-Hydroxypiperidine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Recombinant ReCR
2.2. Rational Design and Characterization of ReCR Variant Y54F
3. Materials and Methods
3.1. Strain and Growth Condition
3.2. Construction, Expression, and Purification of Recombinant Enzyme ReCR
3.3. Enzyme Activity Assays and Characterization of Recombinant ReCR
3.4. Asymmetric Reduction of NBPO Catalyzed by Whole Cells of E. coli BL21(DE3)/pEASY-E2-recr
3.5. Construction, Characterization, and Docking Analysis of ReCR Variant Y54F
3.6. Nucleotide Sequence Accession Number
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Babu, M.S.; Raghunadh, A.; Ramulu, K.; Dahanukar, V.H.; Kumar, U.K.S.; Dubey, P.K. A practical and enantiospecific synthesis of (−)-(R)- and (+)-(S)-piperidin-3-ols. Helv. Chim. Acta 2014, 97, 1507–1515. [Google Scholar] [CrossRef]
- Vitaku, E.; Smith, D.T.; Njardarson, J.T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J. Med. Chem. 2014, 57, 10257–10274. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-F.; Zhang, Y.-P.; Fan, H.-Y.; Wu, K.; Lin, J.-P.; Wang, H.-L.; Wei, D.-Z. Efficient bioreductive production of (R)-N-Boc-3-hydroxypiperidine by a carbonyl reductase. Catal. Commun. 2017, 97, 5–9. [Google Scholar] [CrossRef]
- Ju, X.; Tang, Y.; Liang, X.; Hou, M.; Wan, Z.; Tao, J. Development of a biocatalytic process to prepare (S)-N-Boc-3-hydroxypiperidine. Org. Process Res. Dev. 2014, 18, 827–830. [Google Scholar] [CrossRef]
- Amat, M.; Llor, N.; Huguet, M.; Molins, E.; Espinosa, E.; Bosch, J. Unprecedented oxidation of a phenylglycinol-derived 2-pyridone: Enantioselective synthesis of polyhydroxypiperidines. Org. Lett. 2001, 3, 3257–3260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Zhang, W.-X.; Zheng, G.-W.; Xu, J.-H. Identification of an ε-keto ester reductase for the efficient synthesis of an (R)-α-lipoic acid precursor. Adv. Synth. Catal. 2015, 357, 1697–1702. [Google Scholar] [CrossRef]
- Lacheretz, R.; Pardo, D.G.; Cossy, J. Daucus carota mediated-reduction of cyclic 3-oxo-amines. Org. Lett. 2009, 11, 1245–1248. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-F.; Fan, H.-Y.; Zhang, Y.-P.; Wu, K.; Wang, H.-L.; Lin, J.-P.; Wei, D.-Z. Development of a practical biocatalytic process for (S)-N-Boc-3-hydroxypiperidine synthesis. Tetrahedron Lett. 2017, 58, 1644–1650. [Google Scholar] [CrossRef]
- Xu, G.-P.; Wang, H.-B.; Wu, Z.-L. Efficient bioreductive production of (S)-N-Boc-3-hydroxypiperidine using ketoreductase ChKRED03. Proc. Biochem. 2016, 51, 881–885. [Google Scholar] [CrossRef]
- Hummel, W.; Groger, H. Strategies for regeneration of nicotinamide coenzymes emphasizing self-sufficient closed-loop recycling systems. J. Biotechnol. 2014, 191, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Stamper, W.; Kosjek, B.; Faber, K.; Kroutil, W. Biocatalytic asymmetric hydrogen transfer employing Rhodococcus ruber DSM 44541. J. Org. Chem. 2003, 68, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ma, H.-M.; Yu, H.-L.; Xu, J.-H. Altering the substrate specificity of reductase CgKR1 from Candida glabrata by protein engineering for bioreduction of aromatic α-keto esters. Adv. Synth. Catal. 2014, 356, 1943–1948. [Google Scholar] [CrossRef]
- Turner, N.J.; O’Reilly, E. Biocatalytic retrosynthesis. Nat. Chem. Biol. 2013, 9, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Isotani, K.; Nakamura, M.; Inoue, K.; Isogai, Y.; Makino, Y. Efficient synthesis of optically pure alcohols by asymmetric hydrogen-transfer biocatalysis: Application of engineered enzymes in a 2-propanol-water medium. Appl. Microbiol. Biotechnol. 2012, 93, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Makino, Y.; Inoue, K.; Dairi, T.; Itoh, N. Engineering of phenylacetaldehyde reductase for efficient substrate conversion in concentrated 2-propanol. Appl. Environ. Microbiol. 2005, 71, 4713–4720. [Google Scholar] [CrossRef] [PubMed]
- Au, S.K.; Bommarius, B.R.; Bommarius, A.S. Biphasic reaction system allows for conversion of hydrophobic substrates by amine dehydrogenases. ACS Catal. 2014, 4, 4021–4026. [Google Scholar] [CrossRef]
- De Gonzalo, G.; Lavandera, I.; Faber, K.; Kroutil, W. Enzymatic reduction of ketones in “micro-aqueous” media catalyzed by ADH-A from Rhodococcus ruber. Org. Lett. 2007, 9, 2163–2166. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Matsuda, M.; Mabuchi, M.; Dairi, T.; Wang, J. Chiral alcohol production by NADH-dependent phenylacetaldehyde reductase coupled with in situ regeneration of NADH. Eur. J. Biochem. 2002, 269, 2394–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.-C.; Tang, M.-H.; Ni, Y. Asymmetric synthesis of lipitor chiral intermediate using a robust carbonyl reductase at high substrate to catalyst ratio. J. Mol. Catal. B: Enzym. 2016, 123, 67–72. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Z.-Q.; Lin, C.-P.; Zheng, Y.-G. Efficient biosynthesis of ethyl (R)-4-chloro- 3-hydroxybutyrate using a stereoselective carbonyl reductase from Burkholderia gladioli. BMC Biotechnol. 2016, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Nealon, C.M.; Musa, M.M.; Patel, J.M.; Phillips, R.S. Controlling substrate specificity and stereospecificity of alcohol dehydrogenases. ACS Catal. 2015, 5, 2100–2114. [Google Scholar] [CrossRef]
- Stepankova, V.; Bidmanova, S.; Koudelakova, T.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Strategies for stabilization of enzymes in organic solvents. ACS Catal. 2013, 3, 2823–2836. [Google Scholar] [CrossRef]
- Wang, Z.; Song, Q.; Yu, M.; Wang, Y.; Xiong, B.; Zhang, Y.; Zheng, J.; Ying, X. Characterization of a stereospecific acetoin(diacetyl) reductase from Rhodococcus erythropolis WZ010 and its application for the synthesis of (2S,3S)-2,3-butanediol. Appl. Microbiol. Biotechnol. 2014, 98, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Huang, M.; Song, Q.; Shao, J.; Ying, X. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase from Rhodococcus erythropolis WZ010. Molecules 2015, 20, 7156–7173. [Google Scholar] [CrossRef] [PubMed]
- Abokitse, K.; Hummel, W. Cloning, sequence analysis, and heterologous expression of the gene encoding a (S)-specific alcohol dehydrogenase from Rhodococcus erythropolis DSM 43297. Appl. Microbiol. Biotechnol. 2003, 62, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Morihama, R.; Wang, J.; Okada, K.; Mizuguchi, N. Purification and characterization of phenylacetaldehyde reductase from a styrene-assimilating Corynebacterium strain, ST-10. Appl. Environ. Microbiol. 1997, 63, 3783–3788. [Google Scholar] [PubMed]
- Kasprzak, J.; Bischoff, F.; Rauter, M.; Becher, K.; Baronian, K.; Bode, R.; Schauer, F.; Vorbrodt, H.-M.; Kunze, G. Synthesis of 1-(S)-phenylethanol and ethyl (R)-4-chloro-3-hydroxybutanoate using recombinant Rhodococcus erythropolis alcohol dehydrogenase produced by two yeast species. Biochem. Eng. J. 2016, 106, 107–117. [Google Scholar] [CrossRef]
- Makino, Y.; Dairi, T.; Itoh, N. Engineering the phenylacetaldehyde reductase mutant for improved substrate conversion in the presence of concentrated 2-propanol. Appl. Microbiol. Biotechnol. 2007, 77, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Karabec, M.; Łyskowski, A.; Tauber, K.C.; Steinkellner, G.; Kroutil, W.; Grogan, G.; Gruber, K. Structural insights into substrate specificity and solvent tolerance in alcohol dehydrogenase ADH-A from Rhodococcus ruber DSM 44541. Chem. Commun. 2010, 46, 6314–6316. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Wang, Y.; Xiong, B.; Wu, T.; Xie, L.; Yu, M.; Wang, Z. Characterization of an allylic/benzyl alcohol dehydrogenase from Yokenella sp. strain WZY002, an organism potentially useful for the synthesis of α,β-unsaturated alcohols from allylic aldehydes and ketones. Appl. Environ. Microbiol. 2014, 80, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rojas, E.; Kurt, T.; Schmidt, U.; Meyer, V.; Garbe, L.-A. A bifunctional enzyme from Rhodococcus erythropolis exhibiting secondary alcohol dehydrogenase-catalase activities. Appl. Microbiol. Biotechnol. 2014, 98, 9249–9258. [Google Scholar] [CrossRef] [PubMed]
- Kratzer, R.; Woodley, J.M.; Nidetzky, B. Rules for biocatalyst and reaction engineering to implement effective, NAD(P)H-dependent, whole cell bioreductions. Biotechnol. Adv. 2015, 33, 1641–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glonke, S.; Sadowski, G.; Brandenbusch, C. Applied catastrophic phase inversion: A continuous non-centrifugal phase separation step in biphasic whole-cell biocatalysis. J. Ind. Microbiol. Biotechnol. 2016, 43, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhang, M.; Zhang, X.; Xin, H.; Yang, H. Pickering emulsion as an efficient platform for enzymatic reactions without stirring. ACS Sustain. Chem. Eng. 2016, 4, 6838–6843. [Google Scholar] [CrossRef]
- Yang, C.; Ying, X.; Yu, M.; Zhang, Y.; Xiong, B.; Song, Q.; Wang, Z. Towards the discovery of alcohol dehydrogenases: NAD(P)H fluorescence-based screening and characterization of the newly isolated Rhodococcus erythropolis WZ010 in the preparation of chiral aryl secondary alcohols. J. Ind. Microbiol. Biotechnol. 2012, 39, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |






| Substrate | Relative Activity (%) | Substrate | Relative Activity (%) |
|---|---|---|---|
| N-Boc-3-Piperidone | 100.0 b ± 2.6 | (R/S)-2-Octanol | 100.0 c ± 1.6 |
| 2,3-Butanedione | 189.0 ± 3.4 | (R/S)-2-Pentanol | 61.8 ± 2.3 |
| 2-Octanone | 169.2 ± 2.9 | 2-Propanol | 47.4 ± 0.5 |
| p-Bromoacetophenone | 143.9 ± 4.3 | (R/S)-2-Butanol | 43.8 ± 1.1 |
| Acetoin | 47.2 ± 0.7 | DL-1-Phenylethanol | 31.5 ± 2.1 |
| β-Ionone | 34.8 ± 1.2 | Cyclohexanol | 8.0 ± 1.0 |
| 4-Hydroxy-2-butanone | 31.8 ± 1.1 | 2-Buten-1-ol | 6.8 ± 0.2 |
| 3-Octen-2-one | 25.7 ± 0.7 | (S)-N-Boc-3-Pyrrolidinol | 2.7 ± 0.4 |
| Acetophenone | 25.3 ± 1.0 | (S)-N-Boc-3-Hydroxypiperidine | 0 |
| Hydroxyacetone | 23.6 ± 0.6 | (R)-N-Boc-3-Hydroxypiperidine | 0 |
| N-Boc-3-Pyrrolidone | 9.2 ± 0.5 | ||
| Acetone | 4.8 ± 0.3 | ||
| 2-Bromoacetophenone | 1.8 ± 0.1 |
| Substrate | Coenzyme (mM) | Vmax (U mg−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| NBPO | NADH (0.4) | 103.57 ± 2.46 | 1.74 ± 0.08 | 62.61 ± 1.49 | 35.98 ± 0.86 |
| (S)-NBHP | NAD+ (0.4) | ND b | ND b | ND b | ND b |
| Acetone | NADH (0.4) | 66.30 ± 3.27 | 46.06 ± 2.62 | 40.08 ± 1.98 | 0.87 ± 0.04 |
| 2-Propanol | NAD+ (0.4) | 23.54 ± 0.27 | 1.46 ± 0.06 | 14.22 ± 0.16 | 9.74 ± 0.11 |
| 2-Octanone | NADH (0.4) | 235.54 ± 5.95 | 3.29 ± 0.05 | 142.38 ± 3.11 | 43.28 ± 0.95 |
| (R/S)-2-Octanol | NAD+ (0.4) | 106.57 ± 2.74 | 4.94 ± 0.45 | 64.42 ± 1.66 | 13.04 ± 0.34 |
| Substrate | Coenzyme (mM) | Vmax (U mg−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| NBPO | NADH (0.4) | 140.72 ± 6.52 | 1.73 ± 0.05 | 85.07 ± 3.94 | 49.17 ± 2.28 |
| (S)-NBHP | NAD+ (0.4) | ND b | ND b | ND b | ND b |
| Acetone | NADH (0.4) | 90.46 ± 1.69 | 37.32 ± 0.56 | 54.68 ± 1.02 | 1.47 ± 0.03 |
| 2-Propanol | NAD+ (0.4) | 35.29 ± 0.88 | 1.03 ± 0.05 | 21.32 ± 0.53 | 20.69 ± 0.51 |
| 2-Octanone | NADH (0.4) | 273.75 ± 7.58 | 3.11 ± 0.12 | 165.48 ± 4.58 | 53.21 ± 1.47 |
| (R/S)-2-Octanol | NAD+ (0.4) | 128.19 ± 3.12 | 1.37 ± 0.08 | 77.49 ± 1.88 | 56.56 ± 1.37 |
| Enzyme b | Substrate (M) | Co-substrate (v/v) | Yield (%) | e.e.p (%) c |
|---|---|---|---|---|
| ReCR | NBPO, 0.5 | 2-Propanol, 10% | 72.15 ± 3.51 | >99.9 (S) |
| ReCR Y54F | NBPO, 0.5 | 2-Propanol, 10% | 98.08 ± 1.65 | >99.9 (S) |
| ReCR | NBPO, 1.5 | (R/S)-2-Octanol, 60% | 77.78 ± 2.23 | >99.9 (S) |
| ReCR Y54F | NBPO, 1.5 | (R/S)-2-Octanol, 60% | 95.89 ± 2.37 | >99.9 (S) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, X.; Zhang, J.; Wang, C.; Huang, M.; Ji, Y.; Cheng, F.; Yu, M.; Wang, Z.; Ying, M. Characterization of a Carbonyl Reductase from Rhodococcus erythropolis WZ010 and Its Variant Y54F for Asymmetric Synthesis of (S)-N-Boc-3-Hydroxypiperidine. Molecules 2018, 23, 3117. https://doi.org/10.3390/molecules23123117
Ying X, Zhang J, Wang C, Huang M, Ji Y, Cheng F, Yu M, Wang Z, Ying M. Characterization of a Carbonyl Reductase from Rhodococcus erythropolis WZ010 and Its Variant Y54F for Asymmetric Synthesis of (S)-N-Boc-3-Hydroxypiperidine. Molecules. 2018; 23(12):3117. https://doi.org/10.3390/molecules23123117
Chicago/Turabian StyleYing, Xiangxian, Jie Zhang, Can Wang, Meijuan Huang, Yuting Ji, Feng Cheng, Meilan Yu, Zhao Wang, and Meirong Ying. 2018. "Characterization of a Carbonyl Reductase from Rhodococcus erythropolis WZ010 and Its Variant Y54F for Asymmetric Synthesis of (S)-N-Boc-3-Hydroxypiperidine" Molecules 23, no. 12: 3117. https://doi.org/10.3390/molecules23123117
APA StyleYing, X., Zhang, J., Wang, C., Huang, M., Ji, Y., Cheng, F., Yu, M., Wang, Z., & Ying, M. (2018). Characterization of a Carbonyl Reductase from Rhodococcus erythropolis WZ010 and Its Variant Y54F for Asymmetric Synthesis of (S)-N-Boc-3-Hydroxypiperidine. Molecules, 23(12), 3117. https://doi.org/10.3390/molecules23123117