Enhancing the Bioconversion of Azelaic Acid to Its Derivatives by Response Surface Methodology
Abstract
:1. Introduction
2. Results and Discussion
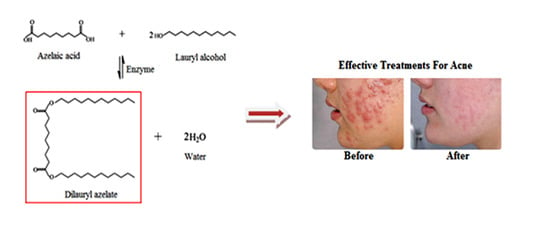
2.1. Identification of Dilauryl Azelate Ester
2.2. Mathematical Model and Analysis of Variance (ANOVA)
2.3. Mutual Effects of Process Parameters
2.4. Reaction Optimization and Model Validation for Dilauryl Azelate Ester
2.5. Evaluation of the Importance of the Variables on the Reaction Conversion
2.6. Antibacterial Assay of Dilauryl Azelate Ester
2.7. Cytotoxicity Assay of Dilauryl Azelate Ester
3. Materials and Methods
3.1. Materials
3.2. Method
3.2.1. Enzymatic Esterification of Azelaic Acid and Analysis of Samples
3.2.2. Antibacterial Assay
3.2.3. Cytotoxicity Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kosmadaki, M.; Katsambas, A. Topical treatments for acne. Clin. Dermatol. 2017, 35, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Khairudin, N.; Basri, M.; Masoumi, H.R.F.; Samiun, W.S.; Samson, S. Lipase-catalyzed synthesis of dilauryl azelate ester: Process optimization by artificial neural networks and reusability study. RSC Adv. 2015, 5, 94909–94918. [Google Scholar] [CrossRef]
- Al-Marabeh, S.; Khalil, E.; Khanfar, M.; Al-Bakri, A.G.; Alzweiri, M. A prodrug approach to enhance azelaic acid percutaneous availability. Pharm. Dev. Technol. 2016, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tamarkin, D. Novel Conjugate Compounds and Dermatological Compositions Thereof. U.S. Patent 20,040,191,196, 30 September 2004. [Google Scholar]
- Manosroi, A.; Panyosak, A.; Rojanasakul, Y.; Manosroi, J. Characteristics and anti-proliferative activity of azelaic acid and its derivatives entrapped in bilayer vesicles in cancer cell lines. J. Drug Target. 2007, 15, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.-Y.; Leu, Y.-L. Prodrug strategy for enhancing drug delivery via skin. Curr. Drug Discov. Technol. 2006, 3, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Svensson, C.K. Biotransformation of drugs in human skin. Drug Metab. Dispos. 2009, 37, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Prausnitz, M.R.; Langer, R. Transdermal drug delivery. Nat. Biotechnol. 2008, 26, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.-W.; Al-Suwayeh, S.A.; Fang, C.-L.; Lin, C.-F.; Chen, C.-C.; Fang, J.-Y. The co-drug of conjugated hydroquinone and azelaic acid to enhance topical skin targeting and decrease penetration through the skin. Eur. J. Pharm. Biopharm. 2012, 81, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Narula, O. Treatise on Fats, Fatty Acids & Oleochemicals; Industrial Consultants: New Delhi, India, 1995. [Google Scholar]
- Cao, L.; Fischer, A.; Bornscheuer, U.T.; Schmid, R.D. Lipase-catalyzed solid phase synthesis of sugar fatty acid esters. Biocatal. Biotransform. 1996, 14, 269–283. [Google Scholar] [CrossRef]
- Chaibakhsh, N.; Abdul Rahman, M.B.; Basri, M.; Salleh, A.B.; Rahman, R.N.Z.R.A. Effect of alcohol chain length on the optimum conditions for lipase-catalyzed synthesis of adipate esters. Biocatal. Biotransform. 2009, 27, 303–308. [Google Scholar] [CrossRef]
- Syamsul, K.; Salina, M.; Siti, S.; Hanina, M.; Basyaruddin, M.; Jusoff, K. Green synthesis of lauryl palmitate via lipase-catalyzed reaction. World Appl. Sci. J. 2010, 11, 401–407. [Google Scholar]
- Zisis, T.; Freddolino, P.L.; Turunen, P.; van Teeseling, M.C.; Rowan, A.E.; Blank, K.G. Interfacial activation of Candida antarctica lipase b: Combined evidence from experiment and simulation. Biochemistry 2015, 54, 5969–5979. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.M.; Larsson, K.M.; Kirk, O. One biocatalyst—Many applications: The use of Candida antarctica b-lipase in organic synthesis. Biocatal. Biotransform. 1998, 16, 181–204. [Google Scholar] [CrossRef]
- Ashari, S.E.; Mohamad, R.; Ariff, A.; Basri, M.; Salleh, A.B. Optimization of enzymatic synthesis of palm-based kojic acid ester using response surface methodology. J. Oleo Sci. 2009, 58, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Khandanlou, R.; Ahmad, M.B.; Masoumi, H.R.F.; Shameli, K.; Basri, M.; Kalantari, K. Rapid adsorption of copper (ii) and lead (ii) by rice straw/fe 3 o 4 nanocomposite: Optimization, equilibrium isotherms, and adsorption kinetics study. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, M.R.; Amiri, S.; Masoumi, H.R.F.; Moghri, M. Optimization of direct yellow 12 dye removal by nanoscale zero-valent iron using response surface methodology. J. Ind. Eng. Chem. 2014, 20, 2535–2542. [Google Scholar] [CrossRef]
- Yetilmezsoy, K.; Demirel, S.; Vanderbei, R.J. Response surface modeling of pb (ii) removal from aqueous solution by Pistacia vera L.: Box—Behnken experimental design. J. Hazard. Mater. 2009, 171, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.H.; Divakar, S.; Prapulla, S.G.; Karanth, N.G. Enzymatic synthesis of isoamyl acetate using immobilized lipase from rhizomucor miehei. J. Biotechnol. 2001, 87, 193–201. [Google Scholar] [CrossRef]
- Radzi, S.M.; Basri, M.; Salleh, A.B.; Ariff, A.; Mohamad, R.; Abdul Rahman, M.B.; Abdul Rahman, R.N.Z. Large scale production of liquid wax ester by immobilized lipase. J. Oleo Sci. 2005, 54, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Basyaruddin, A.R.M.; Chaibakhsh, N.; Basri, M.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Radzi, S.M. Modeling and optimization of lipase-catalyzed synthesis of dilauryl adipate ester by response surface methodology. J. Chem. Technol. Biotechnol. 2008, 83, 1534–1540. [Google Scholar] [Green Version]
- Hari Krishna, S.; Sattur, A.P.; Karanth, N.G. Lipase-catalysed synthesis of isoamly isobutyrate-optimisation using a central composite rotatable design. Process Biochem. 2001, 37, 9–16. [Google Scholar] [CrossRef]
- Masoumi, H.R.F.; Kassim, A.; Basri, M.; Abdullah, D.K.; Haron, M.J. Multivariate optimization in the biosynthesis of a triethanolamine (tea)-based esterquat cationic surfactant using an artificial neural network. Molecules 2011, 16, 5538–5549. [Google Scholar] [CrossRef] [PubMed]
- Ngan, C.L.; Basri, M.; Lye, F.F.; Fard Masoumi, H.R.; Tripathy, M.; Abedi Karjiban, R.; Abdul-Malek, E. Comparison of box—Behnken and central composite designs in optimization of fullerene loaded palm-based nano-emulsions for cosmeceutical application. Ind. Crop Prod. 2014, 59, 309–317. [Google Scholar] [CrossRef]
- Gad, S.C. Alternatives to in vivo studies in toxicology. In General, Applied and Systems Toxicology, 2nd ed.; Ballantyne, B., Marvis, Y., Turner, P., Eds.; Macmillan: New York, NY, USA, 2009. [Google Scholar]
- Chaibakhsh, N.; Rahman, M.B.A.; Abd-Aziz, S.; Basri, M.; Salleh, A.B.; Rahman, R.N.Z.R.A. Optimized lipase-catalyzed synthesis of adipate ester in a solvent-free system. J. Ind. Microbiol. Biotechnol. 2009, 36, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Fard Masoumi, H.R.; Basri, M.; Kassim, A.; Kuang Abdullah, D.; Abdollahi, Y.; Abd Gani, S.S.; Rezaee, M. Statistical optimization of process parameters for lipase-catalyzed synthesis of triethanolamine-based esterquats using response surface methodology in 2-liter bioreactor. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |






| Variables | Units | Coded Level of Variables | |||||
|---|---|---|---|---|---|---|---|
| −2 | −1 | 0 | 1 | 2 | |||
| X1 | Enzyme amount | gram | 0.05 | 0.15 | 0.25 | 0.35 | 0.45 |
| X2 | Reaction time | min | 90 | 180 | 270 | 360 | 450 |
| X3 | Reaction temperature | °C | 40 | 46 | 52 | 58 | 64 |
| X4 | Molar ratio of substrates | AA:LA (mole:mole) | 1:3 | 1:4.5 | 1:6 | 1:7.5 | 1:9 |
| Run No. | Enzyme Amount (g) | Reaction Time (min) | Reaction Temperature (°C) | Molar Ratio of Substrates, AzA:LA (mole) | Degree of Conversion % | |
|---|---|---|---|---|---|---|
| Actual Value | Predicted Value | |||||
| 1 | 0.15 | 180 | 58 | 1:4.5 | 87.83 | 87.82 |
| 2 | 0.15 | 360 | 58 | 1:4.5 | 93.51 | 92.40 |
| 3 | 0.25 | 270 | 52 | 1:6 | 86.61 | 86.24 |
| 4 | 0.35 | 180 | 58 | 1:7.5 | 87.89 | 87.60 |
| 5 | 0.25 | 90 | 52 | 1:6 | 80.80 | 81.19 |
| 6 | 0.35 | 180 | 46 | 1:4.5 | 80.62 | 80.59 |
| 7 | 0.35 | 360 | 58 | 1:4.5 | 95.22 | 96.21 |
| 8 | 0.25 | 270 | 52 | 1:9 | 79.69 | 79.28 |
| 9 | 0.25 | 270 | 52 | 1:6 | 87.95 | 86.24 |
| 10 | 0.35 | 360 | 46 | 1:4.5 | 95.40 | 94.88 |
| 11 | 0.35 | 360 | 46 | 1:7.5 | 90.83 | 89.84 |
| 12 | 0.25 | 450 | 52 | 1:6 | 96.74 | 96.49 |
| 13 | 0.15 | 360 | 58 | 1:7.5 | 86.73 | 87.36 |
| 14 | 0.25 | 270 | 40 | 1:6 | 85.71 | 86.60 |
| 15 | 0.35 | 360 | 58 | 1:7.5 | 90.69 | 91.16 |
| 16 | 0.25 | 270 | 52 | 1:6 | 85.27 | 86.24 |
| 17 | 0.15 | 180 | 46 | 1:7.5 | 83.91 | 82.50 |
| 18 | 0.25 | 270 | 52 | 1:3 | 85.24 | 85.79 |
| 19 | 0.35 | 180 | 46 | 1:7.5 | 78.76 | 79.12 |
| 20 | 0.25 | 270 | 52 | 1:6 | 84.38 | 86.24 |
| 21 | 0.15 | 360 | 46 | 1:4.5 | 95.38 | 95.71 |
| 22 | 0.05 | 270 | 52 | 1:6 | 93.20 | 93.04 |
| 23 | 0.15 | 180 | 58 | 1:7.5 | 84.98 | 86.35 |
| 24 | 0.25 | 270 | 52 | 1:6 | 86.61 | 86.24 |
| 25 | 0.15 | 360 | 46 | 1:7.5 | 90.13 | 90.67 |
| 26 | 0.25 | 270 | 64 | 1:6 | 92.52 | 91.77 |
| 27 | 0.35 | 180 | 58 | 1:4.5 | 89.78 | 89.07 |
| 28 | 0.15 | 180 | 46 | 1:4.5 | 84.14 | 83.96 |
| 29 | 0.25 | 270 | 52 | 1:6 | 86.61 | 86.24 |
| 30 | 0.45 | 270 | 52 | 1:6 | 93.17 | 93.47 |
| Source | Sum of Squares | DF * | Mean Square | F-Value | p-Value Prob > F |
|---|---|---|---|---|---|
| Model | 688.58 | 12 | 57.38 | 51.37 | <0.0001 |
| A-Novozym 435 amount | 0.27 | 1 | 0.27 | 0.24 | 0.6308 |
| B-Reaction time | 351.47 | 1 | 351.47 | 314.67 | <0.0001 |
| C-Reaction temperature | 40.23 | 1 | 40.23 | 36.02 | <0.0001 |
| D-Molar ratio of substrates | 63.52 | 1 | 63.52 | 56.87 | <0.0001 |
| AB | 6.52 | 1 | 6.52 | 5.84 | 0.0272 |
| AC | 21.40 | 1 | 21.40 | 19.16 | 0.0004 |
| BC | 51.28 | 1 | 51.28 | 45.92 | <0.0001 |
| BD | 12.77 | 1 | 12.77 | 11.43 | 0.0035 |
| A2 | 84.45 | 1 | 84.45 | 75.60 | <0.0001 |
| B2 | 11.62 | 1 | 11.62 | 10.40 | 0.0050 |
| C2 | 14.91 | 1 | 14.91 | 13.35 | 0.0020 |
| D2 | 23.47 | 1 | 23.47 | 21.01 | 0.0003 |
| Residual | 18.99 | 17 | 1.12 | - | - |
| Lack of fit | 11.25 | 12 | 0.94 | 0.61 | 0.7794 |
| Pure error | 7.74 | 5 | 1.55 | - | - |
| Standard deviation | 1.06 | ||||
| PRESS | 54.47 | ||||
| R2 | 0.9732 | ||||
| Adjusted R2 | 0.9542 | ||||
| Predicted R2 | 0.9230 | ||||
| Coefficient of variation | 1.20 | ||||
| Adequate Precision | 24.973 |
| Enzyme Amount (gram) | Independent Variables | Conversion% | |||||
|---|---|---|---|---|---|---|---|
| Reaction Time (min) | Reaction Temperature (°C) | Molar Ratio of Substrates, AA:LA (mole) | Actual Value | Predicted Value | RSE (%) | ||
| Validation Set | 0.20 | 180 | 50 | 1:6 | 84.23 | 83.01 | 1.47 |
| 0.30 | 300 | 52 | 1:5 | 91.19 | 89.05 | 2.40 | |
| 0.20 | 360 | 52 | 1:6.5 | 90.99 | 89.84 | 1.28 | |
| 0.16 | 250 | 55 | 1:5.5 | 87.17 | 87.75 | 0.66 | |
| 0.10 | 360 | 52 | 1:6 | 95.10 | 93.54 | 1.66 | |
| Optimal conditions | 0.14 | 360 | 46 | 1:4.1 | 95.38 | 96.23 | 0.88 |
| Samples | Diameter Zone Inhibition (mm)on Bacteria Staphylococcus epidermidis S273 |
|---|---|
| Azelaic acid | 11.5 ± 0.1 mm |
| Dilauryl azelate ester | 9.0 ± 0.1 mm |
| Standard (Streptomycin) | 28.0 ± 0.1 mm |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khairudin, N.; Basri, M.; Fard Masoumi, H.R.; Samson, S.; Ashari, S.E. Enhancing the Bioconversion of Azelaic Acid to Its Derivatives by Response Surface Methodology. Molecules 2018, 23, 397. https://doi.org/10.3390/molecules23020397
Khairudin N, Basri M, Fard Masoumi HR, Samson S, Ashari SE. Enhancing the Bioconversion of Azelaic Acid to Its Derivatives by Response Surface Methodology. Molecules. 2018; 23(2):397. https://doi.org/10.3390/molecules23020397
Chicago/Turabian StyleKhairudin, Nurshafira, Mahiran Basri, Hamid Reza Fard Masoumi, Shazwani Samson, and Siti Efliza Ashari. 2018. "Enhancing the Bioconversion of Azelaic Acid to Its Derivatives by Response Surface Methodology" Molecules 23, no. 2: 397. https://doi.org/10.3390/molecules23020397
APA StyleKhairudin, N., Basri, M., Fard Masoumi, H. R., Samson, S., & Ashari, S. E. (2018). Enhancing the Bioconversion of Azelaic Acid to Its Derivatives by Response Surface Methodology. Molecules, 23(2), 397. https://doi.org/10.3390/molecules23020397