In Silico Discovery of a Substituted 6-Methoxy-quinalidine with Leishmanicidal Activity in Leishmania infantum
Abstract
:1. Introduction
2. Results and Discussion
2.1. Construction of the LiNDH2 Homology Model
2.2. Ligand-Based Pharmacophore Modeling and Virtual Screening
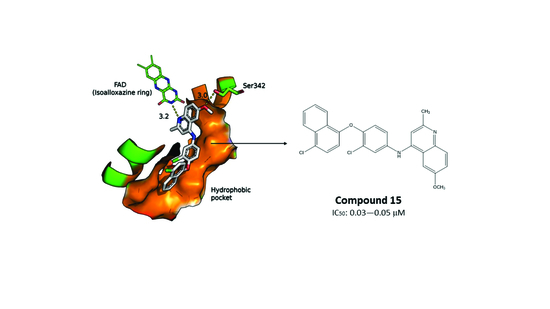
2.3. Molecular Docking of the Pharmacophore-Based Hit Compounds
2.4. Results of the In Vitro Inhibition Assays and Measurements of the Leishmanicidal Effect
3. Materials and Methods
3.1. Construction of the LiNDH2 Homology Model
3.2. Alignment of Protein 3D Structures
3.3. Ligand-Based Pharmacophore Modeling and Virtual Screening
3.4. Molecular Docking Calculations
3.5. Steady-State Kinetics Experiments to Determine Inhibition Activity of NDH-2 from S. aureus
3.6. Determination of Selected Compound Leishmanicidal Effect in Axenic Amastigotes of L. infantum
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CTD | C-terminal domain |
| FAD | Flavin adenine dinucleotide |
| GA | Genetic algorithm |
| HDQ | hydroxy-2-dodecyl-4-(1H) quinolone |
| LiNDH2 | Leishmania infantum NDH-2 |
| NADH | Nicotinamide adenine dinucleotide |
| NDH-2 | Alternative (or type 2) nicotinamide adenine dinucleotide dehydrogenase |
| PfNDH2 | Plasmodium falciparum NDH-2 |
| PAINS | Pan assay interference compounds |
| PDB | Protein data bank |
| RMSD | Root-mean-square deviation |
| ScNDH2 | Saccharomyces cerevisiae NDH-2 |
| SaNDH2 | Staphylococcus aureus NDH-2 |
| UQ | Ubiquinone |
| UQI | 1st Ubiquinone binding site |
| UQII | 2nd Ubiquinone binding site |
References
- WHO. Leishmaniasis: Fact Sheet N°375. 2014. Available online: http://www.who.int/mediacentre/factsheets/fs375/en/ (accessed on 5 February 2018).
- Marreiros, B.C.; Sena, F.P.; Sousa, F.M.; Batista, A.P.; Pereira, M.M. Type II NADH:quinone oxidoreductase family: Phylogenetic distribution, structural diversty and evolutionary divergences. Environ. Microbiol. 2016, 18, 4697–4709. [Google Scholar] [CrossRef] [PubMed]
- Elguindy, M.M.; Nakamaru-Ogiso, E. Apoptosis-iducing Facotr (AIF) and Its Family Member Protein, AMID, Are Rotenone-sensitive NADH: Ubiquinone Oxidoreductases (NDH-2). J. Biol. Chem. 2015, 290, 20815–20826. [Google Scholar] [CrossRef] [PubMed]
- Tomas, A.; Ferreira, C.; Duarte, M. Alternative NADH dehydrogenase of Leishmania: A putative drug target for leishmaniasis. In Proceedings of the COST Action CM1307 2nd Conference, Belgrade, Serbia, 26–28 October 2015; p. 22. [Google Scholar]
- Iwata, M.; Lee, Y.; Yamashita, T.; Yagi, T.; Iwata, S.; Cameron, A.D.; Maher, M.J. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc. Natl. Acad. Sci. USA 2012, 109, 15247–15252. [Google Scholar] [CrossRef] [PubMed]
- Marreiros, B.C.; Sena, F.V.; Sousa, F.M.; Oliveira, A.S.; Soares, C.M.; Batista, A.P.; Pereira, M.M. Structural and Functional insights into the catalytic mechanism of the Type II NADH:quinine oxidoreductase family. Sci. Rep. 2017, 7, 42303. [Google Scholar] [CrossRef] [PubMed]
- Blaza, J.; Bridges, H.; Aragão, D.; Dunn, E.; Heikal, A.; Cook, G.; Nakatani, Y.; Hirst, J. The mechanism of catalysis by type-II NADH:quinone oxidoreductases. Sci. Rep. 2017, 7, 40165. [Google Scholar] [CrossRef] [PubMed]
- Shiba, T.; Kido, Y.; Sakamoto, K.; Inaoka, D.K.; Tsuge, C.; Tatsumi, R.; Takahashi, G.; Balogun, E.O.; Nara, T.; Aoki, T.; et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc. Natl. Acad. Sci. USA 2013, 110, 4580–4585. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Li, W.; Li, J.; Wang, J.; Ge, J.; Xu, D.; Liu, Y.; Wu, K.; Zeng, Q.; Wu, J.W.; et al. Structural insight into type-II mitochondrial NADH dehydrogenases. Nature 2012, 491, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, Y.; Li, X.; Li, J.; Wu, Y.; Yu, J.; Ge, J.; Huang, Z.; Jiang, L.; Rao, Y.; et al. Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule to Eliminate Drug-Resistant Malaria. J. Med. Chem. 2017, 60, 1994–2005. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yamashita, T.; Nakamaru-Ogiso, E.; Hashimoto, T.; Murai, M.; Igarashi, J.; Miyoshi, H.; Mori, N.; Matsuno-Yagi, A.; Yagi, T.; et al. Reaction mechanism of single Subunit NADH-ubiquinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae: Evidence for ternary complex mechanism. J. Biol. Chem. 2011, 286, 9287–9297. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Rahimian, M.; Aneja, K.K.; Schechter, N.M.; Rubin, H.; Scott, C.P. Mycobacterium tuberculosis type II NADH-menaquinone oxidoreductase catalyzes electron transfer through a two-site ping-pong mechanism and has two quinone-binding sites. Biochemistry 2014, 53, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- Heikal, A.; Nakatani, Y.; Dunn, E.; Weimar, M.R.; Day, C.L.; Baker, E.N.; Lott, J.S.; Sazanov, L.A.; Cook, G.M. Structure of the bacterial type II NADH dehydrogenase: A monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 2014, 91, 950–964. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre 2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Sena, F.V.; Batista, A.P.; Catarino, T.; Brito, J.A.; Archer, M.; Viertler, M.; Madl, T.; Cabrita, E.J.; Pereira, M.M. Type-II NADH:quione oxidoreductase from Staphylococcus aureus has two distinct binding sites and is rate limited by quinone reduction. Mol. Microbiol. 2015, 98, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Protein Data Bank. Research Collaboratory for Structural Bioinformatics. Available online: http://www.pdb.org/pdb/home/home.do (accessed on 5 February 2018).
- Chung, S.; Subbiah, S. A structural explanation for twilight zone of protein sequence. Structure 1996, 4, 1123–1127. [Google Scholar] [CrossRef]
- Buchan, D.; Minneci, F.; Nugent, T.; Bryson, K.; Jones, D. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, W349–W357. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.H.; Grishin, N.V. PROMALS3D: A tool for multiple sequence and structure alignment. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [PubMed]
- Vuorinen, A.; Schuster, D. Methods for generating and applying pharmacophore models as virtual screening filters and for bioactivity profiling. Methods 2015, 71, 113–134. [Google Scholar] [CrossRef] [PubMed]
- Biagini, G.A.; Fisher, N.; Shone, A.E.; Mubaraki, M.A.; Srivastava, A.; Hill, A.; Antoine, T.; Warman, A.J.; Davies, J.; Pidathala, C.; et al. Generation of quinolone antimalarials targeting the Plasmodium falciparum mitochondrial respiratory chain for the treatment and prophylaxis of malaria. Proc. Natl. Acad. Sci. USA 2012, 109, 8298–8303. [Google Scholar] [CrossRef] [PubMed]
- Wober, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.; Walters, M.A. Chemistry: Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Gamo, F.; Sanz, L.; Vidal, J.; de Cozar, C.; Alvarez, E.; Lavandera, J.; Vanderwall, D.; Green, D.; Kumar, V.; Hasan, S.; et al. Thousands of chemical starting points for antimalarial lead identification. Nature 2010, 465, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Plouffe, D.; Brinker, A.; McNamara, C.; Henson, K.; Kato, N.; Kuhen, K.; Nagle, A.; Adrián, F.; Matzen, J.; Anderson, P.; et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc. Natl. Acad. Sci. USA 2008, 105, 9059–9064. [Google Scholar] [CrossRef] [PubMed]
- Meister, S.; Plouffe, D.; Kuhen, K.; Bonamy, G.; Wu, T.; Barnes, S.; Bopp, S.; Borboa, R.; Bright, A.; Che, J.; et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science 2011, 334, 1372–1377. [Google Scholar] [CrossRef] [PubMed]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Krüger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Zoidis, G.; Tsotinis, A.; Tsatsaroni, A.; Taylor, M.C.; Kelly, J.M.; Efstathiou, A.; Smirlis, D.; Fytas, G. Lipophilic conformationally constrained spiro carbocyclic 2,6-diketopiperazine-1-acetohydroxamic acid analogues as trypanocidal and leishmanicidal agents: An extended SAR study. Chem. Biol. Drug Des. 2018, 91, 408–421. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., III; De Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; PRichardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins 2002, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Pymol, Version 2.0; The PyMOL Molecular Graphics System; Schrödinger, LLC: New York, NY, USA, 2017.
- ChemDraw Pro Ultra, Version 12.0; Tool for Producing Biological and Chemical Srawings; PerkinElmer: Waltham, MA, USA, 2009.
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.; Carchia, M.; Irwin, J.; Shoichet, B. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Kruber, O. Über das 2.3-Dimethyl-naphthalin im Steinkohlenteer. Eur. J. Inorg. Chem. 1929, 62, 3044–3047. [Google Scholar] [CrossRef]
- Sereno, D.; Lemesre, J.L. Axenically cultured amastigote forms as an in vitro model for investigation of antileishmanial agents. Antimicrob. Agents Chemother. 1997, 41, 972–976. [Google Scholar]
- Vale-Costa, S.; Gomes-Pereira, S.; Teixeira, C.M.; Rosa, G.; Rodrigues, P.N.; Tomas, A.; Appelberg, R.; Gomes, M.S. Iron overload favors the elimination of Leishmania infantum from mouse tissues through interaction with reactive oxygen and nitrogen species. PLoS Negl. Trop Dis. 2013, 7, e2061. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the investigated compounds which were obtained from commercial sources (see Table S3) are available from the authors. |







| Root-Mean-Square Deviation (RMSD) for | Model to 4g73 (No. of Residues and % Aligned) | Model to 4xdb (No. of Residues and % Aligned) |
|---|---|---|
| All atoms | 0.227 Å (321 residues, 74% aligned) | 2.822 Å (370 residues, 85% aligned) |
| Cα-chain | 0.923 Å (396 residues, 91% aligned) | 1.331 Å (252 residues, 58% aligned) |
| Compd. | Chemical Structure | RA | Compd. | Chemical Structure | RA | Compd. | Chemical Structure | RA |
|---|---|---|---|---|---|---|---|---|
| 1 |  | 93% | 9 |  | 80% | 17 |  | 91% |
| 2 |  | 75% | 10 |  | 92% | 18 |  | 86% |
| 3 |  | 71% | 11 |  | 83% | 19 |  | 92% |
| 4 |  | 74% | 12 |  | 95% | 20 |  | 84% |
| 5 |  | 86% | 13 |  | 86% | 21 |  | 94% |
| 6 |  | 89% | 14 |  | 93% | 22 |  | 86% |
| 7 |  | 86% | 15 |  | 49% | 23 |  | 88% |
| 8 |  | 99% | 16 |  | 87% |
| Compound | Wild Type IC50 (µM) | |
|---|---|---|
| Promastigotes | Amastigotes | |
| 11 | 5–10 | >20 |
| 15 | 0.03–0.05 | 0.2–0.3 |
| 20 | 5–10 | >20 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stevanović, S.; Perdih, A.; Senćanski, M.; Glišić, S.; Duarte, M.; Tomás, A.M.; Sena, F.V.; Sousa, F.M.; Pereira, M.M.; Solmajer, T. In Silico Discovery of a Substituted 6-Methoxy-quinalidine with Leishmanicidal Activity in Leishmania infantum. Molecules 2018, 23, 772. https://doi.org/10.3390/molecules23040772
Stevanović S, Perdih A, Senćanski M, Glišić S, Duarte M, Tomás AM, Sena FV, Sousa FM, Pereira MM, Solmajer T. In Silico Discovery of a Substituted 6-Methoxy-quinalidine with Leishmanicidal Activity in Leishmania infantum. Molecules. 2018; 23(4):772. https://doi.org/10.3390/molecules23040772
Chicago/Turabian StyleStevanović, Strahinja, Andrej Perdih, Milan Senćanski, Sanja Glišić, Margarida Duarte, Ana M. Tomás, Filipa V. Sena, Filipe M. Sousa, Manuela M. Pereira, and Tom Solmajer. 2018. "In Silico Discovery of a Substituted 6-Methoxy-quinalidine with Leishmanicidal Activity in Leishmania infantum" Molecules 23, no. 4: 772. https://doi.org/10.3390/molecules23040772
APA StyleStevanović, S., Perdih, A., Senćanski, M., Glišić, S., Duarte, M., Tomás, A. M., Sena, F. V., Sousa, F. M., Pereira, M. M., & Solmajer, T. (2018). In Silico Discovery of a Substituted 6-Methoxy-quinalidine with Leishmanicidal Activity in Leishmania infantum. Molecules, 23(4), 772. https://doi.org/10.3390/molecules23040772