Investigation on Species Authenticity for Herbal Products of Celastrus Orbiculatus and Tripterygum Wilfordii from Markets Using ITS2 Barcoding
Abstract
:1. Introduction
2. Results
2.1. ITS2 Barcode Database for C. orbiculatus, T. wilfordii, and Other Closely Related Species
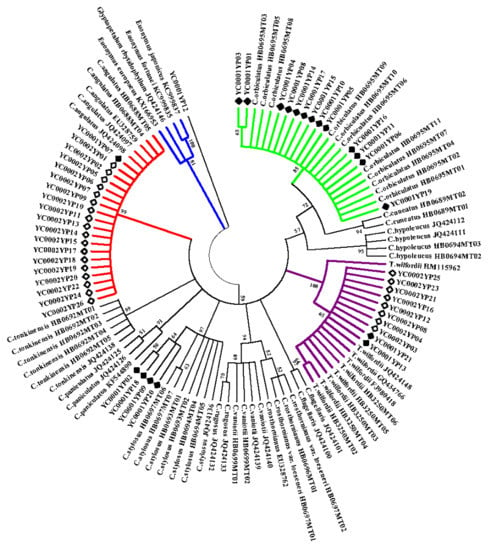
2.2. Identification of Species-Level for C. orbiculatus, T. wilfordii, and Other Closely Related Plants
2.3. Examination of the Species Authenticity for the Herbal Products of C. orbiculatus and T. wilfordii Using ITS2 Barcoding
3. Discussion
3.1. The ITS2 Barcode Can be Used to Identify Celastrus Species and T. wilfordii
3.2. DNA Barcoding Can be Used to Trace Herbal Products of C. orbiculatus and T. wilfordii in Markets
3.3. The Application of DNA Barcoding for Traceability
4. Materials and Methods
4.1. Plant and Herbal Materials
4.2. DNA Extraction
4.3. Amplification, Sequencing, and Sequence Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ma, J.S.; Brach, A.R.; Liu, Q.R. A revision of the genus Tripterygium (Celastraceae). Edinb. J. Bot. 1999, 56, 33–46. [Google Scholar]
- Bao, J.; Dai, S.M. A Chinese herb Tripterygium wilfordii Hook F in the treatment of rheumatoid arthritis: Mechanism, efficacy, and safety. Rheumatol. Int. 2011, 31, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Younger, J.; Fan, F.Z.; Wang, B.; Lipsky, P.E. Benefit of an extract of Tripterygium Wilfordii Hook F in patients with rheumatoid arthritis: A double-blind, placebo-controlled study. Arthrit. Rheumatol. 2002, 46, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Lipsky, P.E.; Tao, X.L. A potential new treatment for rheumatoid arthritis: Thunder god vine. Semin. Arthrit. Rheum. 1997, 26, 713–723. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, Z.; Xiao, J.; Zhang, Y.; Lin, S.; Duan, W.; Yao, J.; Liu, C.; Huang, X.; Wang, T. Effects of triptolide from Tripterygium wilfordii on ERalpha and p53 expression in two human breast cancer cell lines. Phytomed. Int. J. Phytother. Phytopharmacol. 2009, 16, 1006–1013. [Google Scholar]
- Su, X.H.; Zhang, M.L.; Zhan, W.H.; Huo, C.H.; Shi, Q.W.; Gu, Y.C.; Kiyota, H. Chemical and pharmacological studies of the plants from genus Celastrus. Chem. Biodivers. 2009, 6, 146–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, X.; Xiong, X.; Yang, Z.; Sun, Y.; Yang, Z.; Hoffman, R.M.; Liu, Y. Efficacy of the Chinese traditional medicinal herb Celastrus orbiculatus Thunb on human hepatocellular carcinoma in an orthothopic fluorescent nude mouse model. Anticancer Res. 2012, 32, 1213–1220. [Google Scholar] [PubMed]
- Liang, B.Y.; Li, S.Y.; Sun, S.Q. Study of Tripterygium hypoglaucum Hutch and Celastrus orbiculatus thunb by FTIR spectrum. Spectrosc. Spectr. Anal. 2009, 29, 313–317. [Google Scholar]
- Hajibabaei, M.; Singer, G.A.; Hebert, P.D.; Hickey, D.A. DNA barcoding: How it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Schindel, D.E.; Miller, S.E. DNA barcoding a useful tool for taxonomists. Nature 2005, 435, 17. [Google Scholar] [CrossRef] [PubMed]
- Dejean, T.; Valentini, A.; Miquel, C.; Taberlet, P.; Bellemain, E.; Miaud, C. Improved detection of an alien invasive species through environmental DNA barcoding: The example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 2012, 49, 953–959. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Janzen, D.H.; Burns, J.M.; Hallwachs, W.; Hebert, P.D. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. USA 2006, 103, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Resour. 2010, 7, 544–548. [Google Scholar] [CrossRef]
- Parveen, I.; Gafner, S.; Techen, N.; Murch, S.J.; Khan, I.A. DNA Barcoding for the Identification of Botanicals in Herbal Medicine and Dietary Supplements: Strengths and Limitations. Planta Med. 2016, 82, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.K.; Hanner, R.H. DNA barcoding detects market substitution in North American seafood. Food Res. Int. 2008, 41, 828–837. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Q.; Kong, L.; Zheng, X. DNA barcoding and phylogenetic analysis of Pectinidae (Mollusca: Bivalvia) based on mitochondrial COI and 16S rRNA genes. Mol. Biol. Rep. 2011, 38, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, D.; Sarma, R.K.; Shanmughanandhan, D.; Srinivasan, R.; Ramalingam, S. Evaluation of DNA barcode candidates for the discrimination of the large plant family Apocynaceae. Plant Syst. Evol. 2015, 301, 1263–1273. [Google Scholar] [CrossRef]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A renaissance in herbal medicine identification: From morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [PubMed]
- Gu, W.; Song, J.; Cao, Y.; Sun, Q.; Yao, H.; Wu, Q.; Chao, J.; Zhou, J.; Xue, W.; Duan, J. Application of the ITS2 Region for Barcoding Medicinal Plants of Selaginellaceae in Pteridophyta. PLoS ONE 2013, 8, e67818. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Su, Y.; Li, X.; Xue, G.; Wang, Q.; Shi, J.; Wang, L.; Chen, S. Identification of Fritillariae bulbus from adulterants using ITS2 regions. Plant Gene 2016, 7, 42–49. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA Barcodes to Identify Flowering Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [PubMed]
- Simmons, M.P.; Mckenna, M.J.; Bacon, C.D.; Yakobson, K.; Cappa, J.J.; Archer, R.H.; Ford, A.J. Phylogeny of Celastraceae tribe Euonymeae inferred from morphological characters and nuclear and plastid genes. Mol. Phylogenet. Evol. 2012, 62, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Simmons, M.P.; Cappa, J.J.; Archer, R.H.; Ford, A.J.; Eichstedt, D.; Clevinger, C.C. Phylogeny of the Celastreae (Celastraceae) and the relationships of Catha edulis (qat) inferred from morphological characters and nuclear and plastid genes. Mol. Phylogenet. Evol. 2008, 48, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.Y.; Zhao, L.C.; Zhang, Z.X. Phylogeny of Celastrus L. (Celastraceae) inferred from two nuclear and three plastid markers. J. Plant Res. 2012, 125, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Xia, X.; Zhao, L.; Zhang, Z. Celastrus obovatifolius sp. nov. (Celastraceae) from China. Nordic J. Bot. 2012, 30, 53–57. [Google Scholar] [CrossRef]
- Law, S.K.; Simmons, M.P.; Techen, N.; Khan, I.A.; He, M.F.; Shaw, P.C.; But, P.P. Molecular analyses of the Chinese herb Leigongteng (Tripterygium wilfordii Hook.f.). Phytochemistry. 2011, 72, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, M.J.; Lombardi, J.A. Delimitation of the Segregate Genera of Maytenus s. l. (Celastraceae) Based on Morphological and Molecular Characters. Syst. Bot. 2011, 36, 922–932. [Google Scholar] [CrossRef]
- Simmons, M.P.; Clevinger, C.C.; Savolainen, V.; Archer, R.H.; Mathews, S.; Doyle, J.J. Phylogeny of the Celastraceae inferred from phytochrome B gene sequence and morphology. Am. J. Bot. 2001, 88, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Simmons, M.P.; Mckenna, M.J. Phylogeny of Celastraceae Subfamilies Cassinoideae and Tripterygioideae Inferred from Morphological Characters and Nuclear and Plastid Loci. Syst. Bot. 2012, 37, 456–467. [Google Scholar] [CrossRef]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Pang, X.; Liao, B.; Yao, H.; Song, J.; Chen, S. An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci. Rep. 2016, 6, 18723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, N.; Yao, Y.; Liang, X.; Qu, X.; Liu, X.; Zhu, Y.; Yang, D.; Sun, W. Identification of species in Tripterygium (Celastraceae) based on DNA barcoding. Biol. Pharm. Bull. 2016, 39, 1760–1766. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.W.; Ma, S.C.; But, P.P.H.; Mak, T.C.W. Isolation and characterization of spirocaesalmin, a novel rearranged vouacapane diterpenoid from Caesalpinia minax Hance. J. Chem. Soc. Perkin Trans. 2001, 22, 2920–2923. [Google Scholar] [CrossRef]
- Chen, Q.S.; Yuan, J.Q.; Huang, L.N.; Zhuang, Y.; Liu, H.G. Research Progress on Caesalpinia minax. Chin. J. Exp. Tradit. Med. Formulae. 2012, 18, 313–317. [Google Scholar]
- Chen, S.L.; Song, J.Y.; Sun, C.; Xu, J.; Zhu, Y.J.; Verpoorte, R.; Fan, T.P. Herbal genomics: Examining the biology of traditional medicines. Science 2015, 347, S27–S29. [Google Scholar]
- Chen, S.L.; Xu, J.; Liu, C.; Zhu, Y.J.; Nelson, D.R.; Zhou, S.G.; Li, C.F.; Wang, L.Z.; Guo, X.; Sun, Y.Z.; et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 2012, 3, 913. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xu, J.; Xiao, S.; Liao, B.; Gao, Y.; Zhai, C.; Qiu, X.; Wen, X. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. Gigascience 2016, 5, 1–5. [Google Scholar]
- Xu, J.; Chu, Y.; Liao, B.S.; Xiao, S.M.; Yin, Q.G.; Bai, R.; Su, H.; Dong, L.L.; Li, X.W.; Chen, S.L. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascience 2017, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; Andre, C. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Vekemans, X.; Beauwens, T.; Lemaire, M. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.P.; Sarla, N.; Siddiq, E.A. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Li, Q.S.; Li, Y.; Song, J.Y.; Xu, H.B.; Xu, J.; Zhu, Y.J.; Li, X.W.; Gao, H.H.; Dong, L.L.; Qian, J.; et al. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy. New Phytol. 2014, 204, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Li, X.; Yao, H.; Lin, Y.; Ma, X.; Cheng, R.; Song, J.; Ni, L.; Fan, C.; Chen, S. Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci. Rep. 2015, 5, 8337. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Yao, H.; Gao, H.; Zhou, X.; Ma, X.; Xu, C.; Chen, J.; Han, J.; Pang, X.; Xu, R. Super food Lycium barbarum (Solanaceae) traceability via an internal transcribed spacer 2 barcode. Food Res. Int. 2013, 54, 1699–1704. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, M.; Yao, H.; Yang, P.; Xin, T.; Li, B.; Sun, W.; Chen, S. Rapidly discriminate commercial medicinal Pulsatilla chinensis (Bge.) Regel from its adulterants using ITS2 barcoding and specific PCR-RFLP assay. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.Y.; Yao, H.; Luo, K.; Xiang, L.; Ma, X.C.; Han, J.P.; Lin, Y.L.; Song, J.Y.; Chen, S.L. Stability and accuracy of the identification of Notopterygii Rhizoma et Radix using the ITS/ITS2 barcodes. Acta Pharm. Sin. 2012, 47, 1098–1105. [Google Scholar]
- Yang, P.; Li, X.; Zhou, H.; Hu, H.; Zhang, H.; Sun, W.; Wang, Y.; Yao, H. Molecular Identification of Chinese Materia Medica and Its Adulterants Using ITS2 and Barco des: A Case Study on Rhizoma Menispermi. Chin. Med. 2014, 5, 190–198. [Google Scholar] [CrossRef]
- Hou, D.; Song, J.; Shi, L.; Ma, X.; Xin, T.; Han, J.; Xiao, W.; Sun, Z.; Cheng, R.; Yao, H. Stability and accuracy assessment of identification of traditional Chinese materia medica using DNA barcoding: A case study on Flos Lonicerae Japonicae. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Ma, P.; Yao, H.; Song, J.; Chen, K.; Liu, Y. Study on DNA Extraction Method for Chinese Herbs. World Sci. Technol. 2012, 14, 1433–1439. [Google Scholar]
- Pang, X.; Shi, L.; Song, J.; Chen, X.; Chen, S. Use of the potential DNA barcode ITS2 to identify herbal materials. J. Nat. Med. 2013, 67, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liao, B.; Song, J.; Pang, X.; Han, J.; Chen, S. A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene 2013, 530, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Schleicher, T.; Schultz, J.; Müller, T.; Dandekar, T.; Wolf, M. 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene 2009, 430, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S.; Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2014, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Ross, H.A.; Murugan, S.; Li, W.L. Testing the reliability of genetic methods of species identification via simulation. Syst. Biol. 2008, 57, 216–230. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |




| Latin Name | No. of Sequences | Sequence Length (bp) | GC Content (%) | Haplotype No. | No. of Intraspecific Variation Sites |
|---|---|---|---|---|---|
| Celastrus orbiculatus | 11 | 223 | 66.4 | 1 | 0 |
| C. stylosus | 7 | 222–223 | 68.0–68.6 | 2 | 1 |
| C. tonkinensis | 6 | 221 | 66.9 | 1 | 0 |
| C. angulatus | 5 | 223 | 66.8 | 1 | 0 |
| C. hypoleucus | 4 | 221 | 66.5 | 1 | 0 |
| C. vaniotii | 4 | 223 | 66.8 | 1 | 0 |
| C. cuneatus | 2 | 222 | 66.2 | 1 | 0 |
| C. rosthornianus | 2 | 223 | 68.1–68.6 | 2 | 1 |
| C. rosthornianus var. loeseneri | 2 | 223 | 67.7–68.1 | 2 | 1 |
| C. paniculatus | 3 | 223 | 66.3–67.3 | 3 | 2 |
| C. rugosus | 2 | 231 | 66.7 | 1 | 0 |
| C. flagellaris | 2 | 230 | 66.9–67.4 | 2 | 4 |
| Tripterygium wilfordii | 9 | 221–223 | 59.6–60.2 | 2 | 1 |
| Total | 59 | 221–231 | 59.6–68.6 | - | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Hu, X.; Wang, P.; Huang, B.; Sun, W.; Xiong, C.; Hu, Z.; Chen, S. Investigation on Species Authenticity for Herbal Products of Celastrus Orbiculatus and Tripterygum Wilfordii from Markets Using ITS2 Barcoding. Molecules 2018, 23, 967. https://doi.org/10.3390/molecules23040967
Zhang J, Hu X, Wang P, Huang B, Sun W, Xiong C, Hu Z, Chen S. Investigation on Species Authenticity for Herbal Products of Celastrus Orbiculatus and Tripterygum Wilfordii from Markets Using ITS2 Barcoding. Molecules. 2018; 23(4):967. https://doi.org/10.3390/molecules23040967
Chicago/Turabian StyleZhang, Jingjing, Xin Hu, Ping Wang, Bisheng Huang, Wei Sun, Chao Xiong, Zhigang Hu, and Shilin Chen. 2018. "Investigation on Species Authenticity for Herbal Products of Celastrus Orbiculatus and Tripterygum Wilfordii from Markets Using ITS2 Barcoding" Molecules 23, no. 4: 967. https://doi.org/10.3390/molecules23040967
APA StyleZhang, J., Hu, X., Wang, P., Huang, B., Sun, W., Xiong, C., Hu, Z., & Chen, S. (2018). Investigation on Species Authenticity for Herbal Products of Celastrus Orbiculatus and Tripterygum Wilfordii from Markets Using ITS2 Barcoding. Molecules, 23(4), 967. https://doi.org/10.3390/molecules23040967