Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica
Abstract
:1. Introduction
2. Results
2.1. The BBX Gene Family Members in Apple
2.2. Chromosomal Localization, Structure and Multiple Sequence Alignments
2.3. Phylogenetic and Synteny Analysis
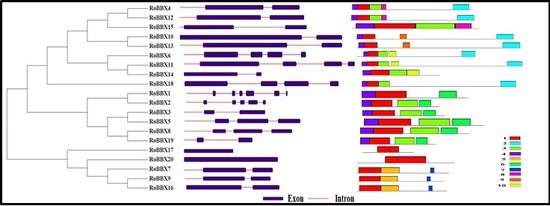
2.4. Gene Structure and Motif Analysis
2.5. Tissue-Specific Analysis of MdBBX Expression Using ArrayExpress Data
2.6. Organ-Specific Expression of BBX Genes in Apple
2.7. Expression of BBX Genes in Response to Flowering Affecting Treatments
3. Materials and Methods
3.1. Identification of BBX Gene Family Member
3.2. Chromosomal Localization and Gene Duplication
3.3. Sequence Alignment and Polygenetic Analysis
3.4. Tandem Duplication and Synteny Analysis
3.5. Plant Materials and Treatments
3.6. Samples Collection
3.7. Quantitative RT-PCR Analysis
4. Discussion
Supplementary Materials
Author Contributions
Conflicts of Interest
Abbreviations
| Ck | control |
| GA | gibberellins; |
| 6-BA | 6-benzylaminopurine; |
| DAFB | days after full bloom |
References
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.-H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- Massiah, M.A.; Matts, J.A.; Short, K.M.; Simmons, B.N.; Singireddy, S.; Yi, Z.; Cox, T.C. Solution structure of the MID1 B-box2 CHC (D/C) C2H2 zinc-binding domain: Insights into an evolutionarily conserved RING fold. J. Mol. Biol. 2007, 369, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Marquardt, K.; Indorf, M.; Jutt, D.; Kircher, S.; Neuhaus, G.; Rodríguez-Franco, M. Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 2011, 156, 1772–1782. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Costa, M.M.R.; Hepworth, S.R.; Vizir, I.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef] [PubMed]
- Ledger, S.; Strayer, C.; Ashton, F.; Kay, S.A.; Putterill, J. Analysis of the function of two circadian—regulated CONSTANS—LIKE genes. Plant J. 2001, 26, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS—LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007, 19, 3242–3255. [Google Scholar] [CrossRef] [PubMed]
- Hassidim, M.; Harir, Y.; Yakir, E.; Kron, I.; Green, R.M. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 2009, 230, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-Y.; Lee, S.-Y.; Seok, H.-Y.; Kim, S.-H.; Sung, Z.R.; Moon, Y.-H. EMF1 interacts with EIP1, EIP6 or EIP9 involved in the regulation of flowering time in Arabidopsis. Plant Cell Physiol. 2011, 52, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Yun, C.-H.; Lee, J.H.; Jang, Y.H.; Park, H.-Y.; Kim, J.-K. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 2008, 228, 355. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Jeong, D.H.; Lee, D.Y.; Yi, J.; Ryu, C.H.; Kim, S.L.; Jeong, H.J.; Choi, S.C.; Jin, P.; Yang, J. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010, 63, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belles-Boix, E.; Babiychuk, E.; Van Montagu, M.; Inzé, D.; Kushnir, S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage 1. FEBS Lett. 2000, 482, 19–24. [Google Scholar] [CrossRef]
- Jaspers, P.; Blomster, T.; Brosche, M.; Salojärvi, J.; Ahlfors, R.; Vainonen, J.P.; Reddy, R.A.; Immink, R.; Angenent, G.; Turck, F. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009, 60, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Fujibe, T.; Saji, H.; Arakawa, K.; Yabe, N.; Takeuchi, Y.; Yamamoto, K.T. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 2004, 134, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tu, X.; Zhang, J.; Chen, X.; Rao, L. Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Boil. Rep. 2013, 40, 2679–2688. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [PubMed]
- Kurokura, T.; Mimida, N.; Battey, N.H.; Hytönen, T. The regulation of seasonal flowering in the Rosaceaee. J. Exp. Bot. 2013, 64, 4131–4141. [Google Scholar] [CrossRef] [PubMed]
- Guitton, B.; Kelner, J.-J.; Velasco, R.; Gardiner, S.E.; Chagné, D.; Costes, E. Genetic control of biennial bearing in apple. J. Exp. Bot. 2011, 63, 131–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, L.-B.; Zhang, D.; Li, Y.-M.; Shen, Y.-W.; Zhao, C.-P.; Ma, J.-J.; An, N.; Han, M.-Y. Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (Malus domestica Borkh.). Plant Cell Physiol. 2015, 56, 2052–2068. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, D.; Lei, C.; Chen, H.; Xing, L.; Ma, J.; Zhao, C.; Han, M. Proteome analyses using iTRAQ labeling reveal critical mechanisms in alternate bearing Malus Prunifolia. J. Proteome Res. 2016, 15, 3602–3616. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, D.; Xing, L.; Zhang, S.; Zhao, C.; Han, M. Effect of exogenous 6-benzylaminopurine (6-BA) on branch type, floral induction and initiation, and related gene expression in ‘Fuji’apple (Malus domestica Borkh.). Plant Growth Regul. 2016, 79, 65–70. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, D.; Fan, S.; Du, L.; Shen, Y.; Xing, L.; Li, Y.; Ma, J.; Han, M. Effect of exogenous GA 3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’apple (Malus domestica Borkh.). Plant Physiol. Biochem. 2016, 107, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Porri, A.; Torti, S.; Romera-Branchat, M.; Coupland, G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 2012, 139, 2198–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Arya, P.; Gupta, K.; Randhawa, V.; Acharya, V.; Singh, A.K. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 2016, 6, 20695. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Chia, T.; Müller, A.; Jung, C.; Mutasa-Göttgens, E. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Experim. Bot. 2008, 59, 2735–2748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34 (Suppl. 2), W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Murelli, C.; Rizza, F.; Albini, F.M.; Dulio, A.; Terzi, V.; Cattivelli, L. Metabolic changes associated with cold-acclimation in contrasting cultivars of barley. Physiol. Plant. 1995, 94, 87–93. [Google Scholar] [CrossRef]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; Sun, Q.; Li, W.; Yu, Y.; Zhao, M.; Meng, Z. Expression analysis of genes encoding double B-box zinc finger proteins in maize. Funct. Integr. Genom. 2017, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Wang, R.; Wang, R.; Yang, S.; Yang, Y. Genome-wide identification, phylogenetic analysis, and expression profiling of the BBX family genes in pear. J. Hort. Sci. Biotechnol. 2017, 93, 1–14. [Google Scholar] [CrossRef]
- Crocco, C.D.; Holm, M.; Yanovsky, M.J.; Botto, J.F. AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 2010, 64, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahn, L.M.; Kong, H.; Leebens-Mack, J.H.; Kim, S.; Soltis, P.S.; Landherr, L.L.; Soltis, D.E.; Ma, H. The evolution of the SEPALLATA subfamily of MADS-box genes. Genetics 2005, 169, 2209–2223. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.Á.; Martínez, M.; Vicente-Carbajosa, J.; Carbonero, P. The family of DOF transcription factors: From green unicellular algae to vascular plants. Mol. Genet. Genom. 2007, 277, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Shigyo, M.; Tabei, N.; Yoneyama, T.; Yanagisawa, S. Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol. 2007, 48, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, D.; Gao, C.; Zhao, M.; Wu, H.; Li, Y.; Shen, Y.; Han, M. Identification, Classification, and Expression Analysis of GRAS Gene Family in Malus domestica. Front. Physiol. 2017, 8, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizukami, Y.; Ma, H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 1997, 9, 393–408. [Google Scholar] [CrossRef] [PubMed]
- Galvão, V.C.; Horrer, D.; Küttner, F.; Schmid, M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 2012. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Xing, L.; Qi, S.; Du, L.; Wu, H.; Shao, H.; Li, Y.; Ma, J.; Han, M. Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus. Mol. Genet. Genom. 2017, 292, 1–17. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |












| S. No. | Sequences | Width | Logo |
|---|---|---|---|
| 1 | HSANKLASRHQRVLLCHVSS | 66 |  |
| 2 | PARVYCKADEAALCVACDAKV | 67 |  |
| 3 | KKTRKFEKTIRYASRKAYAETRPRIKGRF | 21 |  |
| 4 | CDICQEAAAFLFCREDRALLCRECDAIIHAANKRTSKHERF | 16 |  |
| 5 | SPTPWKGSGNKLGPTVSVCESCVNNLEGRHEEDEDEDDDDE | 14 |  |
| 6 | WLIPNPNFNSKLHMDIAPDILKSSDDLIFPEIDSLLEFDYPTSVHTISGS | 5 |  |
| 7 | DDGAEEDEENQVVPLSSTPPPPPPSSSASSREZRLSESVNGDGSSARREP | 8 |  |
| 8 | PLGDGPGIQMPTQLSDMDREARVLRYREK | 15 |  |
| 9 | PVQPDPIPPPSFNMNYNISGPADHNCFDLDFCRSKLYSSFN | 5 |  |
| 10 | MKIQCDSCZKA | 45 |  |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shalmani, A.; Fan, S.; Jia, P.; Li, G.; Muhammad, I.; Li, Y.; Sharif, R.; Dong, F.; Zuo, X.; Li, K.; et al. Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules 2018, 23, 1763. https://doi.org/10.3390/molecules23071763
Shalmani A, Fan S, Jia P, Li G, Muhammad I, Li Y, Sharif R, Dong F, Zuo X, Li K, et al. Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules. 2018; 23(7):1763. https://doi.org/10.3390/molecules23071763
Chicago/Turabian StyleShalmani, Abdullah, Sheng Fan, Peng Jia, Guofang Li, Izhar Muhammad, Youmei Li, Rahat Sharif, Feng Dong, Xiya Zuo, Ke Li, and et al. 2018. "Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica" Molecules 23, no. 7: 1763. https://doi.org/10.3390/molecules23071763
APA StyleShalmani, A., Fan, S., Jia, P., Li, G., Muhammad, I., Li, Y., Sharif, R., Dong, F., Zuo, X., Li, K., Chen, K. -M., & Han, M. (2018). Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules, 23(7), 1763. https://doi.org/10.3390/molecules23071763