Styryl Quinazolinones as Potential Inducers of Myeloid Differentiation via Upregulation of C/EBPα
Abstract
:1. Introduction
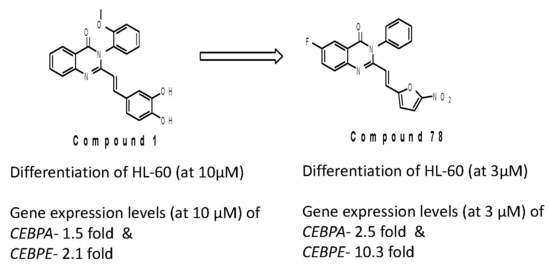
2. Results and Discussion
3. Conclusions
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferrara, F.; Palmieri, S.; Mele, G. Prognostic factors and therapeutic options for relapsed or refractory acute myeloid leukemia. Haematologica 2004, 89, 998–1008. [Google Scholar] [PubMed]
- Zhou, G.B.; Zhang, J.; Wang, Z.Y.; Chen, S.J.; Chen, Z. Treatment of acute promyelocytic leukaemia with all-trans retinoic acid and arsenic trioxide: A paradigm of synergistic molecular targeting therapy. Philos. Trans. R. Soc. B 2007, 362, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; Stewart, D.; Koeffler, H.P. Differentiation therapy of leukemia: 3 decades of development. Blood 2009, 113, 3655–3665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Post, S.M.; Kantarjian, H.; Quintás-Cardama, A. Biology of adult myelocytic leukemia and myelodysplasia. In The Molecular Basis of Cancer, 4th ed.; Elsevier-Saunders: Philadelphia, PA, USA, 2015; pp. 421–432. [Google Scholar]
- Bernlohr, D.A.; Simpson, M.A. Biochemistry of lipids, lipoproteins and membranes. In New Comprehensive Biochemistry; Elsevier: Waltham, MA, USA, 1996; pp. 257–281. [Google Scholar]
- Mcdonald, P.P. Transcriptional Regulation in Neutrophils: Teaching Old Cells New Tricks. Adv. Immun. 2004, 82, 1–48. [Google Scholar] [PubMed]
- Morgan, E.T. Regulation of Drug-Metabolizing Enzymes and Drug Metabolism by Inflammatory Responses. Drug Metab. Dis. 2017, 21–58. [Google Scholar]
- Koleva, R.I.; Ficarro, S.B.; Radomska, H.S.; Carrasco-Alfonso, M.J.; Alberta, J.A.; Webber, J.T.; Luckey, C.J.; Marcucci, G.; Tenen, D.G.; Marto, J.A. C/EBPα and DEK coordinately regulate myeloid differentiation. Blood 2012, 119, 4878–4888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.E.; Zhang, P.; Wang, N.D.; Hetherington, C.J.; Darlington, G.J.; Tenen, D.G. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. USA 1997, 94, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Pabst, T.; Mueller, B.U.; Zhang, P.; Radomska, H.S.; Narravula, S.; Schnittger, S.; Behre, G.; Hiddemann, W.; Tenen, D.G. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat. Genet. 2001, 27, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Koschmieder, S.; Halmos, B.; Levantini, E.; Tenen, D.G. Dysregulation of the C/EBPα Differentiation Pathway in Human Cancer. J. Clin. Oncol. 2009, 27, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Jafari, E.; Khajouei, M.R.; Hassanzadeh, F.; Hakimelahi, G.H.; Khodarahmi, G.A. Quinazolinone and quinazoline derivatives: Recent structures with potent antimicrobial and cytotoxic activities. Res. Pharm. Sci. 2016, 11, 1–14. [Google Scholar] [PubMed]
- Cunningham, D.; Zalcberg, J.; Maroun, J.; James, R.; Clarke, S.; Maughan, T.S.; Vincent, M.; Schulz, J.; Barón, M.G.; Facchini, T. Efficacy, tolerability and management of raltitrexed (Tomudex™) monotherapy in patients with advanced colorectal cancer: A review of phase II/III trials. Eur. J. Cancer 2002, 38, 478–486. [Google Scholar] [CrossRef]
- Penna, L.S.; Henriques, L.A.P.; Bonatto, D. Anti-mitotic agents: Are they emerging molecules for cancer treatment? Pharm. Ther. 2017, 173, 67–82. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, M.J.A.; Dumez, H.; Verweij, J.; Yarkoni, C.; Snyder, D.; Lacombe, D.; Marréaud, S.; Yamaguchi, T.; Punt, C.J.A.; Van Oosterom, A. Phase I and pharmacokinetic study of halofuginone, an oral quinazolinone derivative in patients with advanced solid tumours. Eur. J. Cancer 2006, 42, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Bouley, R.; Kumarasiri, M.; Peng, Z.; Otero, L.H.; Song, W.; Suckow, M.A.; Schroeder, V.A.; Wolter, W.R.; Lastochkin, E.; Antunes, N.T.; et al. Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-cyanostyryl)quinazolin-4(3H)-one. J. Am. Chem. Soc. 2015, 137, 1738–1741. [Google Scholar] [CrossRef] [PubMed]
- Bouley, R.; Ding, D.; Peng, Z.; Bastian, M.; Lastochkin, E.; Song, W.; Suckow, M.A.; Schroeder, V.A.; Wolter, W.R.; Mobashery, S.; et al. Structure–Activity Relationship for the 4(3H)-Quinazolinone Antibacterials. J. Med. Chem. 2016, 59, 5011–5021. [Google Scholar] [CrossRef] [PubMed]
- Raffa, D.; Edler, M.C.; Daidone, G.; Maggio, B.; Merickech, M.; Plescia, S.; Schillaci, D.; Bai, R.; Hamel, E. Synthesis, cytotoxicity, and inhibitory effects on tubulin polymerization of a new 3-heterocyclo substituted 2-styrylquinazolinones. Eur. J. Med. Chem. 2004, 39, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Hour, M.J.; Huang, L.J.; Kuo, S.C.; Xia, Y.; Bastow, K.; Nakanishi, Y.; Hamel, E.; Lee, K.H. 6-Alkylamino- and 2,3-Dihydro-3¢-methoxy-2-phenyl-4-quinazolinones and Related Compounds: Their Synthesis, Cytotoxicity, and Inhibition of Tubulin Polymerization. J. Med. Chem. 2000, 43, 4479–4487. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.B.; Hesson, D.P.; Dusak, B.A.; Dexter, D.L.; Kang, G.J.; Hamel, E. Synthesis and Biological Evaluation of 2-Styrylquinazolin-4(3H)-ones, a New Class of Antimitotic Anticancer Agents Which Inhibit Tubulin Polymerization. J. Med. Chem. 1990, 33, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Sultana, F.; Ramaiah, M.J.; Srikanth, Y.V.V.; Viswanath, A.; Bharathi, E.V.; Nayak, R.; Pushpavalli, S.N.C.V.L.; Srinivas, C.; Pal-Bhadra, M. 3-Diarylethyne quinazolinones: A new class of senescence inducers. Med. Chem. Commun. 2013, 4, 575–581. [Google Scholar] [CrossRef]
- Radomska, H.S.; Jernigan, F.; Nakayama, S.; Jorge, S.E.; Sun, L.; Tenen, D.G.; Kobayashi, S.S. A Cell-Based High-Throughput Screening for Inducers of Myeloid Differentiation. J. Biomol. Screen. 2015, 20, 1150–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compound 78 are available from the authors. |





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sridhar, R.; Takei, H.; Syed, R.; Kobayashi, I.S.; Hui, L.B.; Kamal, A.; Tenen, D.G.; Kobayashi, S.S. Styryl Quinazolinones as Potential Inducers of Myeloid Differentiation via Upregulation of C/EBPα. Molecules 2018, 23, 1938. https://doi.org/10.3390/molecules23081938
Sridhar R, Takei H, Syed R, Kobayashi IS, Hui LB, Kamal A, Tenen DG, Kobayashi SS. Styryl Quinazolinones as Potential Inducers of Myeloid Differentiation via Upregulation of C/EBPα. Molecules. 2018; 23(8):1938. https://doi.org/10.3390/molecules23081938
Chicago/Turabian StyleSridhar, Radhakrishnan, Hisashi Takei, Riyaz Syed, Ikei S. Kobayashi, Liu Bee Hui, Ahmed Kamal, Daniel G. Tenen, and Susumu S. Kobayashi. 2018. "Styryl Quinazolinones as Potential Inducers of Myeloid Differentiation via Upregulation of C/EBPα" Molecules 23, no. 8: 1938. https://doi.org/10.3390/molecules23081938
APA StyleSridhar, R., Takei, H., Syed, R., Kobayashi, I. S., Hui, L. B., Kamal, A., Tenen, D. G., & Kobayashi, S. S. (2018). Styryl Quinazolinones as Potential Inducers of Myeloid Differentiation via Upregulation of C/EBPα. Molecules, 23(8), 1938. https://doi.org/10.3390/molecules23081938