Contributions and Challenges of High Throughput qPCR for Determining Antimicrobial Resistance in the Environment: A Critical Review
Abstract
:1. Introduction
2. Methodology and Meta-Analysis
3. Competitor Technologies of HT-qPCR for High Throughput Profiling of ARGs
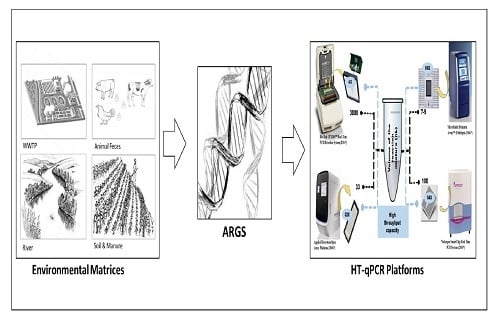
Different HT-qPCR Platforms Used for ARGs Analysis
4. Role and Impact of Community Dynamics and MGEs in ARGs Dissemination
5. Trends of ARGs Diversity in Different Matrices
Potential in Clinical Laboratories
6. Implications of Data Analysis and Quality Control
7. Potential of Other Amplification Technologies for ARGs Measurement
8. Conclusions
Supplementary Materials
Authors Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ciorba, V.; Odone, A.; Veronesi, L.; Pasquarella, C.; Signorelli, C. Antibiotic resistance as a major public health concern: Epidemiology and economic impact. Ann. Ig. 2015, 27, 562–579. [Google Scholar] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar] [PubMed]
- Aubertheau, E.; Stalder, T.; Mondamert, L.; Ploy, M.-C.; Dagot, C.; Labanowski, J. Impact of wastewater treatment plant discharge on the contamination of river biofilms by pharmaceuticals and antibiotic resistance. Sci. Total Environ. 2017, 579, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal pollution explains antibiotic resistance gene abundances in anthropogenically impacted environments. bioRxiv 2018, 341487. [Google Scholar] [CrossRef]
- Larsson, D.G.J. Antibiotics in the environment. Ups. J. Med. Sci. 2014, 119, 108–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aminov, R.I. Horizontal Gene Exchange in Environmental Microbiota. Front. Microbiol. 2011, 2, 158. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Smalla, K. Horizontal gene transfer between bacteria. Environ. Biosafety Res. 2007, 6, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- Suzuki, S.; Ogo, M.; Koike, T.; Takada, H.; Newman, B. Sulfonamide and tetracycline resistance genes in total- and culturable-bacterial assemblages in South African aquatic environments. Front. Microbiol. 2015, 6, 796. [Google Scholar] [CrossRef]
- Jia, S.; He, X.; Bu, Y.; Shi, P.; Miao, Y.; Zhou, H.; Shan, Z.; Zhang, X.-X. Environmental fate of tetracycline resistance genes originating from swine feedlots in river water. J. Environ. Sci. Heal. Part B 2014, 49, 624–631. [Google Scholar] [CrossRef]
- Waseem, H.; Williams, M.R.; Stedtfeld, T.; Chai, B.; Stedtfeld, R.D.; Cole, J.R.; Tiedje, J.M.; Hashsham, S.A. Virulence factor activity relationships (VFARs): A bioinformatics perspective. Environ. Sci. Process. Impacts 2017, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Qian, X.; Gu, J.; Wang, X.-J.; Duan, M.-L. Mechanism and Effect of Temperature on Variations in Antibiotic Resistance Genes during Anaerobic Digestion of Dairy Manure. Sci. Rep. 2016, 6, 30237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Ouyang, W.; Qian, Y.; Su, C.; Su, J.; Chen, H. High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environ. Pollut. 2016, 213, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-L.; An, X.-L.; Zhu, Y.-G.; Su, J.-Q.; Gillings, M.R.; Ye, Z.-L.; Cui, L. Application of Struvite Alters the Antibiotic Resistome in Soil, Rhizosphere and Phyllosphere. Environ. Sci. Technol. 2017, 51, 8149–8157. [Google Scholar] [CrossRef] [PubMed]
- Cui, E.-P.; Gao, F.; Liu, Y.; Fan, X.-Y.; Li, Z.-Y.; Du, Z.-J.; Hu, C.; Neal, A.L. Amendment soil with biochar to control antibiotic resistance genes under unconventional water resources irrigation: Proceed with caution. Environ. Pollut. 2018, 240, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.-H.; Qiao, M.; Su, J.-Q.; Chen, Z.; Zhou, X.; Zhu, Y.-G. High Throughput Profiling of Antibiotic Resistance Genes in Urban Park Soils with Reclaimed Water Irrigation. Environ. Sci. Technol. 2014, 48, 9079–9085. [Google Scholar] [CrossRef] [Green Version]
- De Vries, L.E.; Vallès, Y.; Agersø, Y.; Vaishampayan, P.A.; García-Montaner, A.; Kuehl, J.V.; Christensen, H.; Barlow, M.; Francino, M.P. The Gut as Reservoir of Antibiotic Resistance: Microbial Diversity of Tetracycline Resistance in Mother and Infant. PLoS ONE 2011, 6, e21644. [Google Scholar] [CrossRef]
- Alicea-Serrano, A.M.; Contreras, M.; Magris, M.; Hidalgo, G.; Dominguez-Bello, M.G. Tetracycline resistance genes acquired at birth. Arch. Microbiol. 2013, 195, 447–451. [Google Scholar] [CrossRef]
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Waseem, H.; Williams, M.R.; Stedtfeld, R.D.; Hashsham, S.A. Antimicrobial Resistance in the Environment. Water Environ. Res. 2017, 89, 921–941. [Google Scholar] [CrossRef] [PubMed]
- Case, R.J.; Boucher, Y.; Dahllof, I.; Holmstrom, C.; Doolittle, W.F.; Kjelleberg, S. Use of 16S rRNA and rpoB Genes as Molecular Markers for Microbial Ecology Studies. Appl. Environ. Microbiol. 2007, 73, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Klappenbach, J.A.; Saxman, P.R.; Cole, J.R.; Schmidt, T.M. rrndb: The Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001, 29, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past-Paleontological Statistics. Available online: https://www.uv.es/~pardomv/pe/2001_1/past/pastprog/past (accessed on 1 October 2018).
- Frye, J.G.; Lindsey, R.L.; Rondeau, G.; Porwollik, S.; Long, F.; McClelland, M.; Jackson, C.R.; Englen, M.D.; Meinersmann, R.J.; Berrang, M.E.; et al. Development of a DNA microarray to detect antimicrobial resistance genes identified in the National Center for Biotechnology Information database. Microb. Drug Resist. 2010, 16, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Hu, Y.; Zhu, L.; Yang, X.; Yin, Y.; Lei, F.; Zhu, Y.; Du, Q.; Wang, X.; Meng, Z.; et al. DNA microarray analysis reveals that antibiotic resistance-gene diversity in human gut microbiota is age related. Sci. Rep. 2014, 4, 609. [Google Scholar] [CrossRef]
- Perreten, V.; Vorlet-Fawer, L.; Slickers, P.; Ehricht, R.; Kuhnert, P.; Frey, J. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 2005, 43, 2291–2302. [Google Scholar] [CrossRef]
- Munir, A.; Waseem, H.; Williams, M.R.; Stedtfeld, R.D.; Gulari, E.; Tiedje, J.M.; Hashsham, S.A. Modeling hybridization kinetics of gene probes in a DNA biochip using FEMLAB. Microarrays 2017, 6, 9. [Google Scholar] [CrossRef]
- Jeanty, C.; Longrois, D.; Mertes, P.-M.; Wagner, D.R.; Devaux, Y. An optimized protocol for microarray validation by quantitative PCR using amplified amino allyl labeled RNA. BMC Genomics 2010, 11, 542. [Google Scholar] [CrossRef]
- Morey, J.S.; Ryan, J.C.; Van Dolah, F.M. Microarray validation: Factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online 2006, 8, 175–193. [Google Scholar] [CrossRef]
- Shi, P.; Jia, S.; Zhang, X.X.; Zhang, T.; Cheng, S.; Li, A. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res. 2013, 47, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yang, Y.; Liang, X.; Yu, K.; Zhang, T.; Li, X. Metagenomic Profiles of Antibiotic Resistance Genes (ARGs) between Human Impacted Estuary and Deep Ocean Sediments. Environ. Sci. Technol. 2013, 47, 12753–12760. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Subirats, J.; Sànchez-Melsió, A.; Borrego, C.M.; Balcázar, J.L.; Simonet, P. Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int. J. Antimicrob. Agents 2016, 48, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Pitkänen, L.K.; Sørum, H.; Stedtfeld, R.D.; Tiedje, J.M.; Virta, M. The Resistome of Farmed Fish Feces Contributes to the Enrichment of Antibiotic Resistance Genes in Sediments below Baltic Sea Fish Farms. Front. Microbiol. 2017, 7, 2137. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Pärnänen, K.; Johnson, T.A.; Lyra, C.; Karkman, A.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol. Ecol. 2016, 92, fiw052. [Google Scholar] [CrossRef] [PubMed]
- DiCenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef]
- Brosius, J.; Palmer, M.L.; Kennedy, P.J.; Noller, H.F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1978, 75, 4801–4805. [Google Scholar] [CrossRef]
- Pinard, R.; de Winter, A.; Sarkis, G.J.; Gerstein, M.B.; Tartaro, K.R.; Plant, R.N.; Egholm, M.; Rothberg, J.M.; Leamon, J.H. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics 2006, 7, 216. [Google Scholar] [CrossRef]
- Giardina, E.; Pietrangeli, I.; Martone, C.; Zampatti, S.; Marsala, P.; Gabriele, L.; Ricci, O.; Solla, G.; Asili, P.; Arcudi, G.; et al. Whole genome amplification and real-time PCR in forensic casework. BMC Genomics 2009, 10, 159. [Google Scholar] [CrossRef]
- Sipos, R.; Székely, A.J.; Palatinszky, M.; Révész, S.; Márialigeti, K.; Nikolausz, M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol. Ecol. 2007, 60, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Malhotra, K.; Foltz, L.; Mahoney, W.C.; Schueler, P.A. Interaction and effect of annealing temperature on primers used in differential display RT-PCR. Nucleic Acids Res. 1998, 26, 854–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuang, J.; Yan, X.; Genders, A.J.; Granata, C.; Bishop, D.J. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 2018, 13, e0196438. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-W.; Wang, J.-T.; Li, J.; Li, J.-J.; Ma, Y.-B.; Chen, D.; He, J.-Z. Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils. Environ. Microbiol. 2016, 18, 3896–3909. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-W.; Wang, J.-T.; Li, J.; Shi, X.-Z.; Ma, Y.-B.; Chen, D.; He, J.-Z. Long-Term Nickel Contamination Increases the Occurrence of Antibiotic Resistance Genes in Agricultural Soils. Environ. Sci. Technol. 2017, 51, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Zhang, Q.; Lobos, A.; Senkbeil, J.; Sadowsky, M.J.; Harwood, V.J.; Saeidi, N.; Marinoni, O.; Ishii, S. Precipitation influences pathogenic bacteria and antibiotic resistance gene abundance in storm drain outfalls in coastal sub-tropical waters. Environ. Int. 2018, 116, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, K.D.; Ishii, S.; LaPara, T.M. A Microfluidic Quantitative Polymerase Chain Reaction Method for the Simultaneous Analysis of Dozens of Antibiotic Resistance and Heavy Metal Resistance Genes. Environ. Sci. Technol. Lett. 2018, 5, 20–25. [Google Scholar] [CrossRef]
- Xiong, W.; Sun, Y.; Ding, X.; Wang, M.; Zeng, Z. Selective pressure of antibiotics on ARGs and bacterial communities in manure-polluted freshwater-sediment microcosms. Front. Microbiol. 2015, 6, 194. [Google Scholar] [CrossRef]
- Tian, Z.; Zhang, Y.; Yu, B.; Yang, M. Changes of resistome, mobilome and potential hosts of antibiotic resistance genes during the transformation of anaerobic digestion from mesophilic to thermophilic. Water Res. 2016, 98, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- De Faria, A.C.S.; de Godoy, I.; Sanches, A.A.A.; Iglesias, G.A.; Candido, S.L.; da Paz, R.C.R.; Nakazato, L.; Dutra, V.; de Faria, A.C.S.; de Godoy, I.; et al. Detection of resistance genes and evaluation of water quality at zoo lakes in Brazil. Ciência Rural 2016, 46, 860–866. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Gao, R.; Wei, Y.; Chen, T.; Fan, J.; Zhou, Z.; Makimilua, T.B.; Jiao, Y.; Chen, H. High-throughput profiling and analysis of antibiotic resistance genes in East Tiaoxi River, China. Environ. Pollut. 2017, 230, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Chen, Q.-L.; Zhu, D.; An, X.-L.; Yang, X.-R.; Su, J.-Q.; Qiao, M.; Zhu, Y.-G. Spatial and temporal distribution of antibiotic resistomes in a peri-urban area is associated significantly with anthropogenic activities. Environ. Pollut. 2018, 235, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-H.; Cohen, T.; Grad, Y.H.; Hanage, W.P.; O’Brien, T.F.; Lipsitch, M. Origin and proliferation of multiple-drug resistance in bacterial pathogens. Microbiol. Mol. Biol. Rev. 2015, 79, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Mc Mahon, M.A.S.; Blair, I.S.; Moore, J.E.; Mc Dowell, D.A. The rate of horizontal transmission of antibiotic resistance plasmids is increased in food preservation-stressed bacteria. J. Appl. Microbiol. 2007, 103, 1883–1888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacquiod, S.; Brejnrod, A.; Morberg, S.M.; Abu Al-Soud, W.; Sørensen, S.J.; Riber, L. Deciphering conjugative plasmid permissiveness in wastewater microbiomes. Mol. Ecol. 2017, 26, 3556–3571. [Google Scholar] [CrossRef] [PubMed]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loot, C.; Nivina, A.; Cury, J.; Escudero, J.A.; Ducos-Galand, M.; Bikard, D.; Rocha, E.P.C.; Mazel, D. Differences in Integron Cassette Excision Dynamics Shape a Trade-Off between Evolvability and Genetic Capacitance. MBio 2017, 8, e02296-16. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef]
- Stedtfeld, R.D.; Stedtfeld, T.M.; Waseem, H.; Fitschen-Brown, M.; Guo, X.; Chai, B.; Williams, M.R.; Shook, T.; Logan, A.; Graham, A.; et al. Isothermal assay targeting class 1 integrase gene for environmental surveillance of antibiotic resistance markers. J. Environ. Manage. 2017, 198, 213–220. [Google Scholar] [CrossRef]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation Between Upstream Human Activities and Riverine Antibiotic Resistance Genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef] [PubMed]
- Stedtfeld, R.D.; Williams, M.R.; Fakher, U.; Johnson, T.A.; Stedtfeld, T.M.; Wang, F.; Khalife, W.T.; Hughes, M.; Etchebarne, B.E.; Tiedje, J.M.; et al. Antimicrobial resistance dashboard application for mapping environmental occurrence and resistant pathogens. FEMS Microbiol. Ecol. 2016, 92, fiw020. [Google Scholar] [CrossRef] [PubMed]
- Le, T.-H.; Ng, C.; Chen, H.; Yi, X.Z.; Koh, T.H.; Barkham, T.M.S.; Zhou, Z.; Gin, K.Y.-H. Occurrences and Characterization of Antibiotic Resistant Bacteria and Genetic Determinants of Hospital Wastewaters in a Tropical Country. Antimicrob. Agents Chemother. 2016, 60, 7449–7456. [Google Scholar] [CrossRef] [PubMed]
- Stedtfeld, R.D.; Guo, X.; Stedtfeld, T.M.; Sheng, H.; Williams, M.R.; Hauschild, K.; Gunturu, S.; Tift, L.; Wang, F.; Howe, A.; et al. Primer set 2.0 for highly parallel qPCR array targeting antibiotic resistance genes and mobile genetic elements. FEMS Microbiol. Ecol. 2018, 94, fiy130. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Ju, F.; Cai, L.; Zhang, T. Taxonomic Precision of Different Hypervariable Regions of 16S rRNA Gene and Annotation Methods for Functional Bacterial Groups in Biological Wastewater Treatment. PLoS ONE 2013, 8, e76185. [Google Scholar] [CrossRef]
- Waldron, L.S.; Gillings, M.R. Screening Foodstuffs for Class 1 Integrons and Gene Cassettes. J. Vis. Exp. 2015, 100, e52889. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Dallman, T.; Rubino, S.; Rabsch, W.; Mwaigwisya, S.; Wain, J.; O’Grady, J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2015, 33, 296–300. [Google Scholar] [CrossRef]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 1–6. [Google Scholar] [CrossRef]
- Ouyang, W.-Y.; Huang, F.-Y.; Zhao, Y.; Li, H.; Su, J.-Q. Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl. Microbiol. Biotechnol. 2015, 99, 5697–5707. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.P.; Surette, M.G. Concentration-dependent activity in natural environments. Front. Microbiol. 2013, 4, 20. [Google Scholar] [CrossRef]
- Singh, A.K.; Das, S.; Singh, S.; Gajamer, V.R.; Pradhan, N.; Lepcha, Y.D.; Tiwari, H.K. Prevalence of antibiotic resistance in commensal Escherichia coli among the children in rural hill communities of Northeast India. PLoS ONE 2018, 13, e0199179. [Google Scholar] [CrossRef] [PubMed]
- Hoban, D.; Baquero, F.; Reed, V.; Felmingham, D. Demographic analysis of antimicrobial resistance among Streptococcus pneumoniae: Worldwide results from PROTEKT 1999–2000. Int. J. Infect. Dis. 2005, 9, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Castro-Sánchez, E.; Moore, L.S.P.; Husson, F.; Holmes, A.H. What are the factors driving antimicrobial resistance? Perspectives from a public event in London, England. BMC Infect. Dis. 2016, 16, 465. [Google Scholar] [CrossRef] [PubMed]
- Allcock, S.; Young, E.H.; Holmes, M.; Gurdasani, D.; Dougan, G.; Sandhu, M.S.; Solomon, L.; Török, M.E. Antimicrobial resistance in human populations: Challenges and opportunities. Glob. Heal. Epidemiol. genomics 2017, 2, e4. [Google Scholar] [CrossRef] [PubMed]
- Fahrenfeld, N.; Bisceglia, K.J. Emerging investigators series: Sewer surveillance for monitoring antibiotic use and prevalence of antibiotic resistance: Urban sewer epidemiology. Environ. Sci. Water Res. Technol. 2016, 2, 788–799. [Google Scholar] [CrossRef]
- Townsend, J.P.; Bøhn, T.; Nielsen, K.M. Assessing the probability of detection of horizontal gene transfer events in bacterial populations. Front. Microbiol. 2012, 3, 27. [Google Scholar] [CrossRef]
- Dutta, C.; Pan, A. Horizontal gene transfer and bacterial diversity. J. Biosci. 2002, 27, 27–33. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Skogerbø, G.; Zhang, P.; Chen, R.; He, S.; Huang, D.-W. The human microbiome: A hot spot of microbial horizontal gene transfer. Genomics 2012, 100, 265–270. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Yang, X.; Li, J.; Lv, N.; Liu, F.; Wu, J.; Lin, I.Y.C.; Wu, N.; Weimer, B.C.; Gao, G.F.; et al. The Bacterial Mobile Resistome Transfer Network Connecting the Animal and Human Microbiomes. Appl. Environ. Microbiol. 2016, 82, 6672–6681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, G.T.; Rockweiler, T.J.; Kersey, R.K.; Frye, K.L.; Mitchner, S.R.; Toal, D.R.; Quan, J. Analytical Performance of Multiplexed Screening Test for 10 Antibiotic Resistance Genes from Perianal Swab Samples. Clin. Chem. 2016, 62, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buelow, E.; Bayjanov, J.R.; Majoor, E.; Willems, R.J.; Bonten, M.J.; Schmitt, H.; van Schaik, W. Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol. Ecol. 2018, 94, fiy087. [Google Scholar] [CrossRef] [PubMed]
- Buelow, E.; Bello González, T.D.J.; Fuentes, S.; de Steenhuijsen Piters, W.A.A.; Lahti, L.; Bayjanov, J.R.; Majoor, E.A.M.; Braat, J.C.; van Mourik, M.S.M.; Oostdijk, E.A.N.; et al. Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects. Microbiome 2017, 5, 88. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, G.; Cai, C.; Zhang, J.; Liu, H. Assessing the safety of thermally processed penicillin mycelial dreg following the soil application: Organic matter’s maturation and antibiotic resistance genes. Sci. Total Environ. 2018, 636, 1463–1469. [Google Scholar] [CrossRef]
- Kang, W.; Zhang, Y.-J.; Shi, X.; He, J.-Z.; Hu, H.-W. Short-term copper exposure as a selection pressure for antibiotic resistance and metal resistance in an agricultural soil. Environ. Sci. Pollut. Res. 2018, 25, 29314–29324. [Google Scholar] [CrossRef]
- Stedtfeld, R.D.; Baushke, S.W.; Tourlousse, D.M.; Miller, S.M.; Stedtfeld, T.M.; Gulari, E.; Tiedje, J.M.; Hashsham, S.A. Development and Experimental Validation of a Predictive Threshold Cycle Equation for Quantification of Virulence and Marker Genes by High-Throughput Nanoliter-Volume PCR on the OpenArray Platform. Appl. Environ. Microbiol. 2008, 74, 3831–3838. [Google Scholar] [CrossRef] [Green Version]
- Johnson, T.A.; Stedtfeld, R.D.; Wang, Q.; Cole, J.R.; Hashsham, S.A.; Looft, T.; Zhu, Y.-G.; Tiedje, J.M. Clusters of Antibiotic Resistance Genes Enriched Together Stay Together in Swine Agriculture. MBio 2016, 7, e02214–e02215. [Google Scholar] [CrossRef]
- Stedtfeld, R.D.; Stedtfeld, T.M.; Fader, K.A.; Williams, M.R.; Bhaduri, P.; Quensen, J.; Zacharewski, T.R.; Tiedje, J.M.; Hashsham, S.A. TCDD influences reservoir of antibiotic resistance genes in murine gut microbiome. FEMS Microbiol. Ecol. 2017, 93, fix058. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, M.R.; Stedtfeld, R.D.; Waseem, H.; Stedtfeld, T.; Upham, B.; Khalife, W.; Etchebarne, B.; Hughes, M.; Tiedje, J.M.; Hashsham, S.A. Implications of direct amplification for measuring antimicrobial resistance using point-of-care devices. Anal. Methods 2017, 9, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Kostić, T.; Ellis, M.; Williams, M.R.; Stedtfeld, T.M.; Kaneene, J.B.; Stedtfeld, R.D.; Hashsham, S.A. Thirty-minute screening of antibiotic resistance genes in bacterial isolates with minimal sample preparation in static self-dispensing 64 and 384 assay cards. Appl. Microbiol. Biotechnol. 2015, 99, 7711–7722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, F.; Stedtfeld, R.D.; Waseem, H.; Williams, M.R.; Cupples, A.M.; Tiedje, J.M.; Hashsham, S.A. Most probable number—loop mediated isothermal amplification (MPN-LAMP) for quantifying waterborne pathogens in <25 min. J. Microbiol. Methods 2017, 33, 27–33. [Google Scholar] [CrossRef] [PubMed]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waseem, H.; Jameel, S.; Ali, J.; Saleem Ur Rehman, H.; Tauseef, I.; Farooq, U.; Jamal, A.; Ali, M.I. Contributions and Challenges of High Throughput qPCR for Determining Antimicrobial Resistance in the Environment: A Critical Review. Molecules 2019, 24, 163. https://doi.org/10.3390/molecules24010163
Waseem H, Jameel S, Ali J, Saleem Ur Rehman H, Tauseef I, Farooq U, Jamal A, Ali MI. Contributions and Challenges of High Throughput qPCR for Determining Antimicrobial Resistance in the Environment: A Critical Review. Molecules. 2019; 24(1):163. https://doi.org/10.3390/molecules24010163
Chicago/Turabian StyleWaseem, Hassan, Sana Jameel, Jafar Ali, Hamza Saleem Ur Rehman, Isfahan Tauseef, Uzma Farooq, Asif Jamal, and Muhammad Ishtiaq Ali. 2019. "Contributions and Challenges of High Throughput qPCR for Determining Antimicrobial Resistance in the Environment: A Critical Review" Molecules 24, no. 1: 163. https://doi.org/10.3390/molecules24010163
APA StyleWaseem, H., Jameel, S., Ali, J., Saleem Ur Rehman, H., Tauseef, I., Farooq, U., Jamal, A., & Ali, M. I. (2019). Contributions and Challenges of High Throughput qPCR for Determining Antimicrobial Resistance in the Environment: A Critical Review. Molecules, 24(1), 163. https://doi.org/10.3390/molecules24010163