Fatty Acids-Based Quality Index to Differentiate Worldwide Commercial Pistachio Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Sample Collection
2.3. Preparation of Pistachio Samples
2.4. Chromatographic Conditions
2.5. Multivariate Data Analysis
2.5.1. Principal Component Analysis (PCA)
2.5.2. Linear Discriminant Analysis (LDA)
3. Results and Discussion
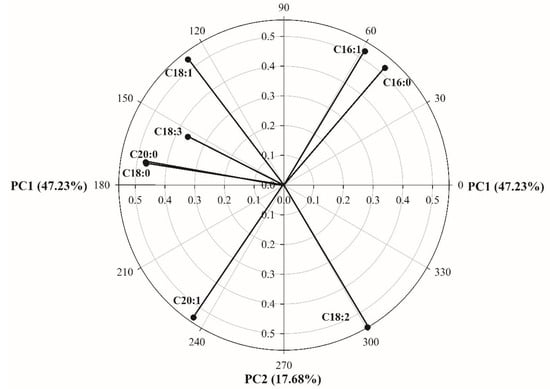
3.1. Unsupervised Pattern Recognition Analysis Using PCA
3.2. Supervised Pattern Recognition Analysis Using LDA
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Azadmard-Damirchi, S.; Emami, S.H.; Hesari, J.; Peighambardoust, S.H.; Nemati, M. Nuts composition and their health benefits. World Acad. Sci. Eng. Technol. 2011, 81, 508–512. [Google Scholar]
- Wang, Q.; Afshin, A.; Yakoob, M.Y.; Singh, G.M.; Rehm, C.D.; Khatibzadeh, S.; Zajkás, G. Impact of nonoptimal intakes of saturated, polyunsaturated, and trans fat on global burdens of coronary heart disease. J. Am. Heart Assoc. 2016. [Google Scholar] [CrossRef] [PubMed]
- Arena, E.; Campisi, S.; Fallico, B.; Maccarone, E. Distribution of fatty acids and phytosterols as a criterion to discriminate geographic origin of pistachio seeds. Food Chem. 2007, 104, 403–408. [Google Scholar] [CrossRef]
- Kasliwal, R.R.; Bansal, M.; Mehrotra, R.; Yeptho, K.P.; Trehan, N. Effect of pistachio nut consumption on endothelial function and arterial stiffness. Nutrition 2015, 31, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Kocyigit, A.; Koylu, A.A.; Keles, H. Effects of pistachio nuts consumption on plasma lipid profile and oxidative status in healthy volunteers. Nutr. Metab. Cardiovasc. Dis. 2006, 16, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Dreher, M.L. Pistachio nuts: Composition and potential health benefits. Nut. Rev. 2012, 70, 234–240. [Google Scholar] [CrossRef]
- Sheridan, M.J.; Cooper, J.N.; Erario, M.; Cheifetz, C.E. Pistachio nut consumption and serum lipid levels. J. Am. Coll. Nutr. 2007, 26, 141–148. [Google Scholar] [CrossRef]
- Aceña, L.; Vera, L.; Guasch, J.; Busto, O.; Mestres, M. Comparative study of two extraction techniques to obtain representative aroma extracts for being analysed by gas chromatography-olfactometry: Application to roasted pistachio aroma. J. Chromatogr. A 2010, 1217, 7781–7787. [Google Scholar] [CrossRef]
- Behgar, M.; Ghasemi, S.; Naserian, A.; Borzoie, A.; Fatollahi, H. Gamma radiation effects on phenolics, antioxidants activity and in vitro digestion of pistachio (Pistachia vera) hull. Radiat. Phys. Chem. 2011, 80, 963–967. [Google Scholar] [CrossRef]
- Sonmezdag, A.S.; Kelebek, H.; Selli, S. Characterization and comparative evaluation of volatile, phenolic and antioxidant properties of pistachio (Pistacia vera L.) hull. J. Essent. Oil Res. 2017, 29, 262–270. [Google Scholar] [CrossRef]
- Fani, S.R.; Zamanizadeh, H.R.; Mirabolfathy, M. Isolation and identification of the causal agents of root and crown rot of pistachio trees in the Sistan and Baluchistan provinces. Acta Horticulturae 2006, 726, 647–650. [Google Scholar] [CrossRef]
- Kouchakzadeh, A.; Tavakoli, T. New practical method for evaluation of a conventional flat plate continuous pistachio dryer. Energ. Convers. Manag. 2011, 52, 2735–2740. [Google Scholar] [CrossRef]
- Baghizadeh, A.; Haghi, R. Studying Genetic Diversity of Pistachio Cultivars in Kerman Province Based on Morphological Traits Using Fourier Series and Cluster Analysis Studying Genetic Diversity of Pistachio Cultivars in Kerman Province Based on Morphological Traits Using Fourier series And Cluster Analysis Studying Genetic Diversity of Pistachio Cultivars in Kerman Province Based on Morphological Traits Using Fourier Series and Cluster Analysis. Glob. J. Sci. Front. Res. Bio-Tech Genet. 2012, 12, 1–5. [Google Scholar]
- Pazouki, L.; Mardi, M.; Shanjani, P.S.; Hagidimitriou, M.; Pirseyedi, S.M.; Naghavi, M.R.; Khayam Nekoui, S.M. Genetic diversity and relationships among Pistacia species and cultivars. Conserv. Genet. 2010, 11, 311–318. [Google Scholar] [CrossRef]
- Esteki, M.; Farajmand, B.; Amanifar, S.; Barkhordari, R.; Ahadiyan, Z.; Dashtaki, E.; Vander Heyden, Y. Classification and authentication of Iranian walnuts according to their geographical origin based on gas chromatographic fatty acid fingerprint analysis using pattern recognition methods. Chemom. Intell. Lab. Syst. 2017, 171, 251–258. [Google Scholar] [CrossRef]
- Esteki, M.; Farajmand, B.; Kolahderazi, Y.; Simal-Gandara, J. Chromatographic Fingerprinting with Multivariate Data Analysis for Detection and Quantification of Apricot Kernel in Almond Powder. Food Anal. Methods 2017, 10, 3312–3320. [Google Scholar] [CrossRef]
- Esteki, M.; Vander Heyden, Y.; Farajmand, B.; Kolahderazi, Y. Qualitative and quantitative analysis of peanut adulteration in almond powder samples using multi-elemental fingerprinting combined with multivariate data analysis methods. Food Control 2017, 82, 31–41. [Google Scholar] [CrossRef]
- Esteki, M.; Shahsavari, Z.; Simal-Gandara, J. Use of spectroscopic methods in combination with linear discriminant analysis for authentication of food products. Food Control 2018, 91, 100–112. [Google Scholar] [CrossRef]
- Esteki, M.; Simal-Gandara, J.; Shahsavari, Z.; Zandbaaf, S.; Dashtaki, E.; Vander Heyden, Y. A review on the application of chromatographic methods, coupled to chemometrics, for food authentication. Food Control 2018, 93, 165–182. [Google Scholar] [CrossRef]
- Esteki, M.; Regueiro, J.; Simal-Gandara, J. Tackling fraudsters with global strategies to reveal fraud in the food chain. Compr. Rev. Food Sci. Food Saf. 2018, in press. [Google Scholar]
- Gonzalez-Fernandez, I.; Iglesias-Otero, M.A.; Moldes, O.A.; Mejuto, J.C.; Simal-Gandara, J.; Esteki, M. A critical review of the use of artificial neural networks with olive oil production and characterization technologies. Crit. Rev. Food Sci. Nutr. 2017, 57, 2896–2908. [Google Scholar]
- Dyszel, S.M.; Pettit, B.C. Determination of the country of origin of pistachio nuts by DSC and HPLC. J. Am. Oil Chem. Soc. 1990, 67, 947–951. [Google Scholar] [CrossRef]
- Anderson, K.A.; Smith, B.W. Effect of season and variety on the differentiation of geographic growing origin of pistachios by stable isotope profiling. J. Agric. Food Chem. 2006, 54, 1747–1752. [Google Scholar] [CrossRef] [PubMed]
- Zur, K.; Heier, A.; Blaas, K.W.; Fauhl-Hassek, C. Authenticity control of pistachios based on 1H- and 13C-NMR spectroscopy and multivariate statistics. Eur. Food Res. Technol. 2008, 227, 969–977. [Google Scholar] [CrossRef]
- Kouchakzadeh, A.; Brati, A. Discrimination of Pistachios Varieties with Neural Network using some Physical Characteristic. Int. J. Emerg. Sci. 2012, 2, 259–268. [Google Scholar]
- Omid, M.; Mahmoudi, A.; Aghagolzadeh, A.; Borghaee, A.M. Seperating pistachio varieties using automatic trainable classifier. In Proceedings of the 3rd International Conference on Autonomic and Autonomous Systems, ICAS’07, Athens, Greece, 9–25 June 2007. [Google Scholar]
- Brereton, R.G. Pattern recognition in chemometrics. Chemom. Intell. Lab. Syst. 2015, 149, 90–96. [Google Scholar] [CrossRef]
- Berente, B.; de La Calle García, D.; Reichenbächer, M.; Danzer, K. Method development for the determination of anthocyanins in red wines by high-performance liquid chromatography and classification of German red wines by means of multivariate statistical methods. J. Chromatogr. A 2000, 871, 95–103. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007. [Google Scholar] [CrossRef]
- Vandeginste, B.G.M.; Massart, M.D.L. Handbook of Chemometrics and Qualimetrics; Elsevier: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Cavazza, A.; Corradini, C.; Musci, M.; Salvadeo, P. High-performance liquid chromatographic phenolic compound fingerprint for authenticity assessment of honey. J. Sci. Food Agric. 2013, 93, 1169–1175. [Google Scholar] [CrossRef]
- Toher, D.; Downey, G.; Murphy, T.B. A comparison of model-based and regression classification techniques applied to near infrared spectroscopic data in food authentication studies. Chemom. Intell. Lab. Syst. 2007, 89, 102–115. [Google Scholar] [CrossRef]
- Yiyu, C.; Minjun, C.; Welsh, W.J. Fractal Fingerprinting of Chromatographic Profiles Based on Wavelet Analysis and Its Application to Characterize the Quality Grade of Medicinal Herbs. J. Chem. Inf. Comput. Sci. 2003, 43, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Lu, W.; Cai, Z.; Bao, L.; Hartmann, C.; Gao, B.; Yu, L. Rapid detection of milk adulteration using intact protein flow injection mass spectrometric fingerprints combined with chemometrics. Food Chem. 2018, 240, 573–578. [Google Scholar] [CrossRef]
- Pepi, S.; Sardella, A.; Bonazza, A.; Vaccaro, C. Geochemical caper fingerprints as a tool for geographical origin identification. Environ. Geochem. Health 2018, 40, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi-Varnamkhasti, M. Sensory stability of pistachio nut (Pistacia vera L.) varieties during storage using descriptive analysis combined with chemometrics. Eng. Agric. Environ. Food 2015, 8, 106–113. [Google Scholar] [CrossRef]
- Hung, Y.-C.; Chen, P.; Chen, L.-Y. Advanced classification of coffee beans with fatty acids profiling to block information loss. Symmetry 2018, 10, 529–539. [Google Scholar] [CrossRef]
- Satil, F.; Azcan, N.; Baser, K.H.C. Fatty acid composition of pistachio nuts in Turkey. Chem. Nat. Compd. 2003. [Google Scholar] [CrossRef]
- Mazinani, S.; Elhami Rad, A.H.; Khaneghah, A.M. Determination and comparison of the amount of tocopherolic and phenolic compounds and fatty acids profile in edible nuts (Pistachio, Almond and Walnut) oil. Adv. Environ. Biol. 2012, 6, 1610–1619. [Google Scholar]
- Davis, J.P.; Dean, L.O.; Faircloth, W.H.; Sanders, T.H. Physical and chemical characterizations of normal and high-oleic oils from nine commercial cultivars of peanut. JAOCS J. Am. Oil Chem. Soc. 2008, 85, 235–243. [Google Scholar] [CrossRef]
- Roozban, M.R.; Mohamadi, N.; Vahdati, K. Fat content and fatty acid composition of four Iranian pistachio (Pistacia vera L.) varieties grown in Iran; IV International Symposium on Pistachios and Almonds: Tehran, Iran, 2006; Volume 726, pp. 573–577. [Google Scholar]
- Casini, C.; Dardanelli, J.L.; Martínez, M.J.; Balzarini, M.; Borgogno, C.S.; Nassetta, M. Oil quality and sugar content of peanuts (Arachis hypogaea) grown in Argentina: Their relationship with climatic variables and seed yield. J. Agric. Food Chem. 2003, 51, 6309–6313. [Google Scholar] [CrossRef]
- Garrett, D.; Peterson, D.A.; Anderson, C.W.; Thaut, M.H. Comparison of linear, nonlinear, and feature selection methods for EEG signal classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2003, 11, 141–144. [Google Scholar] [CrossRef]
Sample Availability: Samples of Pistachio are available from the authors. |



| Number of Sample | Pistachio Cultivar | Fatty Acid Composition (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C16:0 (Palmitic) | C16:1 (Palmitoleic) | C18:0 (Stearic) | C18:1 (Oleic) | C18:2 (Linoleic) | C18:3 (Linolenic) | C20:0 (Arachidic) | C20:1 (Gondoic) | ||
| 1 | Ahm | 8.89 | 0.53 | 1.52 | 64.59 | 23.50 | 0.29 | 0.16 | 0.52 |
| 2 | Ahm | 8.87 | 0.52 | 1.51 | 64.51 | 23.61 | 0.28 | 0.17 | 0.53 |
| 3 | Ahm | 8.91 | 0.55 | 1.45 | 63.55 | 24.56 | 0.29 | 0.15 | 0.54 |
| 4 | Ahm | 8.82 | 0.52 | 1.52 | 64.55 | 23.6 | 0.29 | 0.17 | 0.53 |
| 5 | Ahm | 8.61 | 0.54 | 1.47 | 62.73 | 25.66 | 0.30 | 0.16 | 0.53 |
| 6 | Ahm | 8.90 | 0.53 | 1.53 | 64.43 | 23.64 | 0.29 | 0.16 | 0.52 |
| 7 | Ahm | 8.65 | 0.55 | 1.39 | 62.02 | 26.43 | 0.28 | 0.14 | 0.54 |
| 8 | Ahm | 8.67 | 0.58 | 1.32 | 61.40 | 27.03 | 0.30 | 0.15 | 0.55 |
| 9 | Ahm | 8.75 | 0.53 | 1.33 | 64.04 | 24.33 | 0.31 | 0.16 | 0.55 |
| 10 | Ahm | 8.78 | 0.58 | 1.26 | 61.33 | 27.07 | 0.30 | 0.14 | 0.54 |
| 11 | Ahm | 8.81 | 0.52 | 1.53 | 64.54 | 23.61 | 0.28 | 0.17 | 0.54 |
| 12 | Ahm | 8.66 | 0.54 | 1.50 | 63.46 | 24.85 | 0.29 | 0.16 | 0.54 |
| 13 | Ahm | 8.89 | 0.52 | 1.55 | 64.57 | 23.50 | 0.28 | 0.16 | 0.53 |
| 14 | Ahm | 8.77 | 0.53 | 1.43 | 62.28 | 26.03 | 0.28 | 0.16 | 0.52 |
| 15 | Ahm | 8.91 | 0.52 | 1.52 | 64.59 | 23.48 | 0.28 | 0.17 | 0.53 |
| 16 | Ahm | 8.61 | 0.53 | 1.44 | 63.11 | 25.31 | 0.29 | 0.16 | 0.55 |
| 17 | Ahm | 8.89 | 0.52 | 1.53 | 64.62 | 23.46 | 0.28 | 0.17 | 0.53 |
| 18 | Ahm | 8.66 | 0.54 | 1.51 | 64.15 | 24.15 | 0.29 | 0.16 | 0.54 |
| 19 | Ahm | 8.88 | 0.57 | 1.35 | 61.29 | 26.89 | 0.30 | 0.15 | 0.57 |
| 20 | Ahm | 8.79 | 0.53 | 1.38 | 63.09 | 25.19 | 0.31 | 0.15 | 0.56 |
| 21 | Ahm | 8.82 | 0.59 | 1.42 | 65.10 | 23.08 | 0.32 | 0.16 | 0.51 |
| 22 | Ahm | 8.69 | 0.58 | 1.39 | 64.12 | 24.20 | 0.31 | 0.15 | 0.56 |
| 23 | Ahm | 8.86 | 0.59 | 1.35 | 65.08 | 23.13 | 0.32 | 0.16 | 0.51 |
| 24 | Ahm | 8.72 | 0.57 | 1.42 | 65.14 | 23.18 | 0.29 | 0.16 | 0.52 |
| 25 | Ahm | 8.86 | 0.59 | 1.37 | 65.09 | 23.10 | 0.32 | 0.16 | 0.51 |
| 26 | Ahm | 8.73 | 0.58 | 1.33 | 64.26 | 24.14 | 0.28 | 0.15 | 0.53 |
| 27 | Ahm | 8.88 | 0.59 | 1.38 | 65.02 | 23.14 | 0.32 | 0.16 | 0.51 |
| 28 | Ahm | 8.63 | 0.56 | 1.38 | 64.28 | 24.16 | 0.28 | 0.15 | 0.56 |
| 29 | Ahm | 8.59 | 0.54 | 1.46 | 63.03 | 25.41 | 0.28 | 0.16 | 0.53 |
| 30 | Ahm | 8.65 | 0.54 | 1.29 | 61.64 | 26.88 | 0.29 | 0.17 | 0.54 |
| 31 | Akb | 8.37 | 0.58 | 1.51 | 69.63 | 18.91 | 0.29 | 0.16 | 0.55 |
| 32 | Akb | 8.56 | 0.57 | 1.45 | 69.45 | 18.95 | 0.31 | 0.17 | 0.54 |
| 33 | Akb | 8.38 | 0.57 | 1.51 | 69.67 | 18.86 | 0.30 | 0.16 | 0.55 |
| 34 | Akb | 8.52 | 0.59 | 1.53 | 68.99 | 19.38 | 0.28 | 0.17 | 0.54 |
| 35 | Akb | 8.38 | 0.57 | 1.52 | 69.65 | 18.86 | 0.30 | 0.16 | 0.56 |
| 36 | Akb | 8.43 | 0.58 | 1.48 | 69.55 | 18.95 | 0.29 | 0.18 | 0.54 |
| 37 | Akb | 8.40 | 0.57 | 1.52 | 69.62 | 18.88 | 0.30 | 0.16 | 0.55 |
| 38 | Akb | 8.59 | 0.58 | 1.49 | 69.77 | 18.57 | 0.30 | 0.16 | 0.54 |
| 39 | Akb | 8.35 | 0.56 | 1.52 | 69.70 | 18.86 | 0.29 | 0.16 | 0.56 |
| 40 | Akb | 8.51 | 0.56 | 1.50 | 68.95 | 19.49 | 0.28 | 0.17 | 0.54 |
| 41 | Akb | 8.75 | 0.59 | 1.44 | 68.57 | 19.69 | 0.28 | 0.16 | 0.52 |
| 42 | Akb | 8.61 | 0.58 | 1.52 | 69.15 | 19.14 | 0.29 | 0.17 | 0.54 |
| 43 | Akb | 8.74 | 0.59 | 1.45 | 68.59 | 19.67 | 0.28 | 0.16 | 0.52 |
| 44 | Akb | 8.42 | 0.58 | 1.53 | 69.66 | 18.82 | 0.30 | 0.16 | 0.53 |
| 45 | Akb | 8.67 | 0.59 | 1.44 | 68.68 | 19.66 | 0.28 | 0.16 | 0.52 |
| 46 | Akb | 8.55 | 0.57 | 1.49 | 69.26 | 19.11 | 0.29 | 0.18 | 0.55 |
| 47 | Akb | 8.33 | 0.56 | 1.54 | 69.82 | 18.72 | 0.29 | 0.17 | 0.57 |
| 48 | Akb | 8.45 | 0.59 | 1.51 | 68.95 | 19.50 | 0.28 | 0.16 | 0.56 |
| 49 | Akb | 8.40 | 0.57 | 1.53 | 69.71 | 18.79 | 0.29 | 0.16 | 0.55 |
| 50 | Akb | 8.99 | 0.58 | 1.54 | 69.61 | 18.29 | 0.28 | 0.17 | 0.54 |
| 51 | Akb | 9.08 | 0.60 | 1.52 | 68.71 | 19.05 | 0.31 | 0.19 | 0.54 |
| 52 | Akb | 8.51 | 0.57 | 1.53 | 68.88 | 19.48 | 0.30 | 0.16 | 0.57 |
| 53 | Akb | 8.70 | 0.58 | 1.49 | 69.84 | 18.37 | 0.30 | 0.18 | 0.54 |
| 54 | Akb | 8.66 | 0.57 | 1.50 | 69.25 | 19.03 | 0.30 | 0.16 | 0.53 |
| 55 | Akb | 8.96 | 0.60 | 1.57 | 69.23 | 18.63 | 0.30 | 0.18 | 0.53 |
| 56 | Akb | 8.48 | 0.59 | 1.54 | 69.62 | 18.73 | 0.29 | 0.17 | 0.58 |
| 57 | Akb | 8.90 | 0.60 | 1.56 | 69.32 | 18.61 | 0.30 | 0.18 | 0.53 |
| 58 | Akb | 8.52 | 0.58 | 1.55 | 69.87 | 18.46 | 0.29 | 0.16 | 0.57 |
| 59 | Akb | 8.41 | 0.56 | 1.56 | 69.42 | 18.98 | 0.30 | 0.18 | 0.59 |
| 60 | Akb | 8.55 | 0.58 | 1.56 | 68.85 | 19.41 | 0.31 | 0.16 | 0.58 |
| 61 | Chr | 9.40 | 0.64 | 1.36 | 64.39 | 23.26 | 0.31 | 0.15 | 0.48 |
| 62 | Chr | 9.46 | 0.57 | 1.55 | 65.16 | 22.35 | 0.28 | 0.15 | 0.48 |
| 63 | Chr | 9.63 | 0.66 | 1.31 | 66.22 | 23.25 | 0.31 | 0.15 | 0.47 |
| 64 | Chr | 9.23 | 0.56 | 1.54 | 65.33 | 22.41 | 0.29 | 0.16 | 0.48 |
| 65 | Chr | 9.16 | 0.54 | 1.50 | 66.84 | 21.00 | 0.28 | 0.17 | 0.51 |
| 66 | Chr | 9.15 | 0.59 | 1.44 | 66.66 | 21.23 | 0.29 | 0.15 | 0.49 |
| 67 | Chr | 9.21 | 0.55 | 1.53 | 66.75 | 21.02 | 0.28 | 0.16 | 0.50 |
| 68 | Chr | 9.13 | 0.53 | 1.52 | 65.70 | 22.21 | 0.28 | 0.15 | 0.48 |
| 69 | Chr | 9.19 | 0.55 | 1.56 | 66.78 | 20.97 | 0.28 | 0.16 | 0.51 |
| 70 | Chr | 9.26 | 0.54 | 1.49 | 66.59 | 21.13 | 0.30 | 0.17 | 0.52 |
| 71 | Chr | 9.18 | 0.54 | 1.47 | 66.85 | 21.02 | 0.28 | 0.16 | 0.50 |
| 72 | Chr | 9.46 | 0.53 | 1.56 | 65.08 | 22.41 | 0.29 | 0.18 | 0.49 |
| 73 | Chr | 9.06 | 0.54 | 1.75 | 64.98 | 22.66 | 0.30 | 0.17 | 0.54 |
| 74 | Chr | 9.08 | 0.54 | 1.56 | 64.78 | 23.06 | 0.30 | 0.15 | 0.53 |
| 75 | Chr | 9.11 | 0.54 | 1.63 | 64.99 | 22.73 | 0.30 | 0.17 | 0.53 |
| 76 | Chr | 9.51 | 0.57 | 1.50 | 66.03 | 21.42 | 0.30 | 0.15 | 0.52 |
| 77 | Chr | 9.23 | 0.55 | 1.49 | 66.68 | 21.09 | 0.29 | 0.16 | 0.51 |
| 78 | Chr | 9.13 | 0.59 | 1.62 | 67.37 | 20.35 | 0.28 | 0.17 | 0.49 |
| 79 | Chr | 9.20 | 0.55 | 1.50 | 66.70 | 21.09 | 0.29 | 0.16 | 0.51 |
| 80 | Chr | 9.51 | 0.61 | 1.49 | 66.67 | 20.75 | 0.28 | 0.15 | 0.54 |
| 81 | Chr | 9.27 | 0.56 | 1.44 | 66.62 | 21.18 | 0.29 | 0.15 | 0.49 |
| 82 | Chr | 9.37 | 0.60 | 1.54 | 67.04 | 20.54 | 0.28 | 0.15 | 0.48 |
| 83 | Chr | 9.19 | 0.56 | 1.69 | 67.45 | 20.10 | 0.31 | 0.19 | 0.51 |
| 84 | Chr | 9.16 | 0.51 | 1.65 | 66.21 | 21.48 | 0.29 | 0.18 | 0.52 |
| 85 | Chr | 9.21 | 0.57 | 1.80 | 67.34 | 20.08 | 0.31 | 0.19 | 0.50 |
| 86 | Chr | 8.67 | 0.51 | 1.61 | 65.93 | 22.32 | 0.29 | 0.15 | 0.52 |
| 87 | Chr | 8.43 | 0.50 | 1.78 | 65.55 | 22.66 | 0.30 | 0.19 | 0.59 |
| 88 | Chr | 9.36 | 0.52 | 1.59 | 65.54 | 22.05 | 0.28 | 0.15 | 0.51 |
| 89 | Chr | 9.03 | 0.53 | 1.56 | 65.21 | 22.67 | 0.30 | 0.17 | 0.53 |
| 90 | Chr | 9.46 | 0.58 | 1.60 | 66.13 | 21.32 | 0.28 | 0.16 | 0.51 |
| 91 | Kal | 10.10 | 0.79 | 1.37 | 66.90 | 19.96 | 0.28 | 0.16 | 0.44 |
| 92 | Kal | 9.33 | 0.75 | 1.28 | 66.78 | 21.03 | 0.27 | 0.13 | 0.43 |
| 93 | Kal | 10.12 | 0.79 | 1.36 | 66.96 | 19.93 | 0.27 | 0.15 | 0.42 |
| 94 | Kal | 9.66 | 0.74 | 1.30 | 66.28 | 21.17 | 0.28 | 0.12 | 0.45 |
| 95 | Kal | 10.53 | 0.88 | 1.32 | 66.61 | 19.82 | 0.29 | 0.14 | 0.41 |
| 96 | Kal | 9.67 | 0.76 | 1.43 | 64.84 | 22.45 | 0.27 | 0.12 | 0.46 |
| 97 | Kal | 9.78 | 0.77 | 1.45 | 67.11 | 19.99 | 0.29 | 0.16 | 0.45 |
| 98 | Kal | 10.02 | 0.79 | 1.40 | 63.75 | 23.20 | 0.27 | 0.15 | 0.42 |
| 99 | Kal | 10.09 | 0.78 | 1.41 | 67.40 | 19.47 | 0.26 | 0.16 | 0.43 |
| 100 | Kal | 9.65 | 0.69 | 1.35 | 67.19 | 20.33 | 0.26 | 0.12 | 0.41 |
| 101 | Kal | 10.09 | 0.78 | 1.42 | 67.51 | 19.35 | 0.26 | 0.16 | 0.43 |
| 102 | Kal | 9.32 | 0.75 | 1.36 | 68.57 | 19.15 | 0.27 | 0.12 | 0.46 |
| 103 | Kal | 10.15 | 0.77 | 1.39 | 67.43 | 19.43 | 0.26 | 0.15 | 0.42 |
| 104 | Kal | 9.15 | 0.68 | 1.30 | 62.17 | 25.75 | 0.27 | 0.15 | 0.53 |
| 105 | Kal | 9.19 | 0.73 | 1.21 | 64.29 | 23.68 | 0.27 | 0.16 | 0.47 |
| 106 | Kal | 9.05 | 0.67 | 1.31 | 62.85 | 25.18 | 0.26 | 0.15 | 0.53 |
| 107 | Kal | 9.66 | 0.71 | 1.21 | 64.94 | 22.58 | 0.26 | 0.14 | 0.50 |
| 108 | Kal | 9.14 | 0.68 | 1.31 | 62.09 | 25.83 | 0.26 | 0.15 | 0.54 |
| 109 | Kal | 10.15 | 0.69 | 1.32 | 64.33 | 22.64 | 0.27 | 0.14 | 0.46 |
| 110 | Kal | 9.55 | 0.74 | 1.00 | 65.89 | 21.94 | 0.28 | 0.13 | 0.47 |
| 111 | Kal | 10.23 | 0.81 | 1.12 | 64.96 | 22.02 | 0.26 | 0.14 | 0.46 |
| 112 | Kal | 9.57 | 0.74 | 0.99 | 65.91 | 21.91 | 0.28 | 0.13 | 0.47 |
| 113 | Kal | 10.10 | 0.79 | 1.43 | 65.45 | 21.37 | 0.27 | 0.14 | 0.45 |
| 114 | Kal | 9.57 | 0.74 | 0.98 | 65.98 | 21.91 | 0.28 | 0.12 | 0.42 |
| 115 | Kal | 9.76 | 0.80 | 1.42 | 63.84 | 23.26 | 0.27 | 0.15 | 0.50 |
| 116 | Kal | 9.01 | 0.65 | 1.29 | 62.39 | 25.69 | 0.26 | 0.16 | 0.55 |
| 117 | Kal | 9.33 | 0.75 | 1.15 | 64.82 | 23.03 | 0.26 | 0.14 | 0.52 |
| 118 | Kal | 9.17 | 0.66 | 1.31 | 62.18 | 25.72 | 0.26 | 0.15 | 0.55 |
| 119 | Kal | 9.26 | 0.71 | 1.09 | 64.68 | 23.32 | 0.27 | 0.16 | 0.51 |
| 120 | Kal | 9.31 | 0.73 | 0.99 | 64.40 | 23.71 | 0.27 | 0.13 | 0.46 |
| 121 | Oha | 8.99 | 0.54 | 1.25 | 61.42 | 26.87 | 0.26 | 0.15 | 0.52 |
| 122 | Oha | 9.05 | 0.46 | 1.22 | 63.79 | 24.54 | 0.26 | 0.16 | 0.52 |
| 123 | Oha | 8.96 | 0.54 | 1.29 | 61.42 | 26.85 | 0.26 | 0.15 | 0.53 |
| 124 | Oha | 9.15 | 0.47 | 1.30 | 61.66 | 26.46 | 0.27 | 0.15 | 0.54 |
| 125 | Oha | 8.92 | 0.53 | 1.27 | 61.52 | 26.82 | 0.26 | 0.15 | 0.53 |
| 126 | Oha | 9.01 | 0.58 | 1.35 | 63.58 | 24.55 | 0.26 | 0.16 | 0.51 |
| 127 | Oha | 9.36 | 0.45 | 1.21 | 65.28 | 22.77 | 0.27 | 0.15 | 0.51 |
| 128 | Oha | 9.05 | 0.41 | 1.22 | 63.93 | 24.44 | 0.26 | 0.17 | 0.52 |
| 129 | Oha | 8.89 | 0.52 | 1.25 | 65.61 | 22.79 | 0.26 | 0.15 | 0.53 |
| 130 | Oha | 9.10 | 0.58 | 1.22 | 61.28 | 26.89 | 0.26 | 0.15 | 0.52 |
| 131 | Oha | 8.86 | 0.53 | 1.41 | 61.48 | 26.77 | 0.26 | 0.15 | 0.54 |
| 132 | Oha | 8.88 | 0.51 | 1.31 | 61.36 | 27.01 | 0.26 | 0.15 | 0.52 |
| 133 | Oha | 8.87 | 0.53 | 1.32 | 61.56 | 26.77 | 0.26 | 0.15 | 0.54 |
| 134 | Oha | 9.00 | 0.49 | 1.45 | 62.00 | 26.13 | 0.27 | 0.15 | 0.51 |
| 135 | Oha | 8.95 | 0.53 | 1.23 | 61.49 | 26.87 | 0.26 | 0.15 | 0.52 |
| 136 | Oha | 8.99 | 0.49 | 1.43 | 64.00 | 24.14 | 0.26 | 0.16 | 0.53 |
| 137 | Oha | 8.94 | 0.53 | 1.40 | 61.41 | 26.79 | 0.26 | 0.15 | 0.52 |
| 138 | Oha | 8.86 | 0.51 | 1.23 | 61.18 | 27.26 | 0.27 | 0.17 | 0.52 |
| 139 | Oha | 8.94 | 0.53 | 1.26 | 61.51 | 26.83 | 0.26 | 0.15 | 0.52 |
| 140 | Oha | 9.06 | 0.59 | 1.32 | 61.97 | 26.12 | 0.26 | 0.15 | 0.53 |
| 141 | Oha | 8.97 | 0.53 | 1.30 | 61.41 | 26.86 | 0.26 | 0.15 | 0.52 |
| 142 | Oha | 9.01 | 0.60 | 1.28 | 62.17 | 26.01 | 0.26 | 0.16 | 0.51 |
| 143 | Oha | 8.96 | 0.53 | 1.24 | 61.46 | 26.88 | 0.26 | 0.15 | 0.52 |
| 144 | Oha | 8.91 | 0.46 | 1.30 | 61.41 | 26.98 | 0.27 | 0.15 | 0.52 |
| 145 | Oha | 8.96 | 0.53 | 1.32 | 61.37 | 26.88 | 0.26 | 0.15 | 0.53 |
| 146 | Oha | 9.03 | 0.52 | 1.28 | 62.67 | 25.57 | 0.26 | 0.16 | 0.51 |
| 147 | Oha | 9.14 | 0.54 | 1.26 | 60.72 | 27.39 | 0.26 | 0.15 | 0.54 |
| 148 | Oha | 8.87 | 0.59 | 1.44 | 63.05 | 25.12 | 0.26 | 0.15 | 0.52 |
| 149 | Oha | 9.22 | 0.60 | 1.49 | 65.33 | 22.43 | 0.26 | 0.17 | 0.50 |
| 150 | Oha | 9.08 | 0.45 | 1.25 | 62.11 | 26.18 | 0.26 | 0.15 | 0.52 |
| Pistachio Cultivar | Ahm | Akb | Chr | Kal | Oha | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Range | Average | Range | Average | Range | Average | Range | Average | Range | Average | |
| C16:0 (Palmitic) | 8.58–8.91 | 8.77 | 8.35–9.08 | 8.57 | 8.43–9.63 | 9.23 | 9.01–10.53 | 9.66 | 8.86–9.36 | 8.99 |
| C16:1 (Palmitoleic) | 0.52–0.59 | 0.55 | 0.56–0.6 | 0.58 | 0.50–0.66 | 0.55 | 0.65–0.88 | 0.74 | 0.45–0.6 | 0.52 |
| C18:0 (Stearic) | 1.26–1.55 | 1.43 | 1.44–1.57 | 1.51 | 1.31–1.80 | 1.55 | 0.98–1.45 | 1.28 | 1.21–1.49 | 1.30 |
| C18:1 (Oleic) | 61.38–65.09 | 63.72 | 68.57–69.84 | 69.34 | 64.39–67.46 | 66.05 | 62.09–67.43 | 65.28 | 60.71–65.29 | 62.30 |
| C18:2 (Linoleic) | 23.08–27.07 | 24.54 | 18.37–19.68 | 18.99 | 20.08–23.26 | 21.65 | 19.47–25.83 | 22.16 | 22.43–27.39 | 25.9 |
| C18:3 (Linolenic) | 0.28–0.32 | 0.29 | 0.28–0.31 | 0.29 | 0.28–0.31 | 0.29 | 0.26–0.29 | 0.27 | 0.26–0.27 | 0.26 |
| C20:0 (Arachidic) | 0.14–0.17 | 0.16 | 0.16–0.19 | 0.16 | 0.15–0.19 | 0.16 | 0.12–0.16 | 0.14 | 0.15–0.17 | 0.15 |
| C20:1 (Gondoic) | 0.51–0.57 | 0.54 | 0.52–0.59 | 0.54 | 0.47–0.59 | 0.51 | 0.41–0.55 | 0.47 | 0.50–0.54 | 0.52 |
| Quality Index | 2.60 | 3.66 | 3.05 | 2.94 | 2.40 | |||||
| Fatty Acid | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | C18:3 | C20:0 | C20:1 |
|---|---|---|---|---|---|---|---|---|
| C16:0 | 1 | |||||||
| C16:1 | 0.745 | 1 | ||||||
| C18:0 | −0.312 | −0.365 | 1 | |||||
| C18:1 | −0.039 | 0.209 | 0.477 | 1 | ||||
| C18:2 | −0.109 | −0.315 | −0.462 | −0.988 | 1 | |||
| C18:3 | −0.247 | −0.064 | 0.419 | 0.46 | −0.433 | 1 | ||
| C20:0 | −0.304 | −0.338 | 0.905 | 0.47 | −0.454 | 0.406 | 1 | |
| C20:1 | −0.921 | −0.730 | 0.3 | −0.112 | 0.241 | 0.112 | 0.341 | 1 |
| Group Predicted | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Original Group | Number of Cases | Ahm | Akb | Chr | Kal | Oha | Accuracy (%) | Sensitivity | Specificity | |
| Ahm | 10 | 10 | 0 | 0 | 0 | 0 | 100 | 100 | 98 | |
| Akb | 10 | 0 | 10 | 0 | 0 | 0 | 100 | 100 | 100 | |
| Chr | 10 | 1 | 0 | 9 | 0 | 0 | 90 | 90 | 100 | |
| Kal | 10 | 0 | 0 | 0 | 10 | 0 | 100 | 100 | 100 | |
| Oha | 10 | 0 | 0 | 0 | 0 | 10 | 100 | 100 | 100 | |
| Predictive ability | 98.0 | 98.0 | 99.6 | |||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esteki, M.; Ahmadi, P.; Vander Heyden, Y.; Simal-Gandara, J. Fatty Acids-Based Quality Index to Differentiate Worldwide Commercial Pistachio Cultivars. Molecules 2019, 24, 58. https://doi.org/10.3390/molecules24010058
Esteki M, Ahmadi P, Vander Heyden Y, Simal-Gandara J. Fatty Acids-Based Quality Index to Differentiate Worldwide Commercial Pistachio Cultivars. Molecules. 2019; 24(1):58. https://doi.org/10.3390/molecules24010058
Chicago/Turabian StyleEsteki, Mahnaz, Parvin Ahmadi, Yvan Vander Heyden, and Jesus Simal-Gandara. 2019. "Fatty Acids-Based Quality Index to Differentiate Worldwide Commercial Pistachio Cultivars" Molecules 24, no. 1: 58. https://doi.org/10.3390/molecules24010058
APA StyleEsteki, M., Ahmadi, P., Vander Heyden, Y., & Simal-Gandara, J. (2019). Fatty Acids-Based Quality Index to Differentiate Worldwide Commercial Pistachio Cultivars. Molecules, 24(1), 58. https://doi.org/10.3390/molecules24010058