Bacterial Lipid II Analogs: Novel In Vitro Substrates for Mammalian Oligosaccharyl Diphosphodolichol Diphosphatase (DLODP) Activities
Abstract
:1. Introduction
2. Results
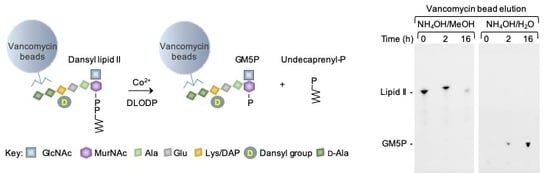
2.1. Generation of Vancomycin Affinity Beads for Solid-Phase Extraction of d-Ala–d-Ala-Containing Compounds
2.2. Bacterial Lipid II is Hydrolyzed by Mouse Liver Microsomal Proteins to Yield GlcNAcβ1,4MurNAc(pentapeptide)-P
2.3. Bacterial Lipid I and Short-Chain Lipid II Analogs Are Also Hydrolyzed by Liver Microsomal Proteins
2.4. The Biochemical Characteristics of GM5P Generation by Liver Microsomal Protein Extracts Are Similar to Those of the Mammalian DLODP Activity
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Synthesis of Bacterial Lipids and Their Precursors and Dolichol-Linked Oligosaccharide Analogs
4.3. Liver Microsomes and Protein Solubilization
4.4. DLODP Assay Using [3H]Man5GlcNAc2-PP-dolichol-2
4.5. Preparation of Vancomycin Beads
4.6. Assay of Bacterial Lipid II Hydrolysis
4.7. Digestion of Bacterial Lipids with Colicin M
4.8. Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Haltiwanger, R.S.; Lowe, J.B. Role of glycosylation in development. Annu. Rev. Biochem. 2004, 73, 491–537. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.B.; Marth, J.D. A genetic approach to Mammalian glycan function. Annu. Rev. Biochem. 2003, 72, 643–691. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.G.; Freeze, H.H. Perspectives on Glycosylation and Its Congenital Disorders. Trends Genet. 2018, 34, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Péanne, R.; de Lonlay, P.; Foulquier, F.; Kornak, U.; Lefeber, D.J.; Morava, E.; Pérez, B.; Seta, N.; Thiel, C.; Van Schaftingen, E.; et al. Congenital disorders of glycosylation (CDG): Quo vadis? Eur. J. Med. Genet. 2018, 61, 643–663. [Google Scholar] [CrossRef] [PubMed]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 2013, 1833, 2430–2437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornfeld, R.; Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar] [CrossRef] [PubMed]
- Chantret, I.; Dupré, T.; Delenda, C.; Bucher, S.; Dancourt, J.; Barnier, A.; Charollais, A.; Heron, D.; Bader-Meunier, B.; Danos, O.; et al. Congenital disorders of glycosylation type Ig is defined by a deficiency in dolichyl-P-mannose: Man7GlcNAc2-PP-dolichyl mannosyltransferase. J. Biol. Chem. 2002, 277, 25815–25822. [Google Scholar] [CrossRef] [PubMed]
- Peric, D.; Durrant-Arico, C.; Delenda, C.; Dupré, T.; De Lonlay, P.; de Baulny, H.O.; Pelatan, C.; Bader-Meunier, B.; Danos, O.; Chantret, I.; et al. The compartmentalisation of phosphorylated free oligosaccharides in cells from a CDG Ig patient reveals a novel ER-to-cytosol translocation process. PLoS ONE 2010, 5, e11675. [Google Scholar] [CrossRef] [PubMed]
- Vleugels, W.; Duvet, S.; Peanne, R.; Mir, A.-M.; Cacan, R.; Michalski, J.-C.; Matthijs, G.; Foulquier, F. Identification of phosphorylated oligosaccharides in cells of patients with a congenital disorders of glycosylation (CDG-I). Biochimie 2011, 93, 823–833. [Google Scholar] [CrossRef]
- Harada, Y.; Nakajima, K.; Masahara-Negishi, Y.; Freeze, H.H.; Angata, T.; Taniguchi, N.; Suzuki, T. Metabolically programmed quality control system for dolichol-linked oligosaccharides. Proc. Natl. Acad. Sci. USA 2013, 110, 19366–19371. [Google Scholar] [CrossRef] [Green Version]
- Aebi, M.; Hennet, T. Congenital disorders of glycosylation: Genetic model systems lead the way. Trends Cell Biol. 2001, 11, 136–141. [Google Scholar] [CrossRef]
- Belard, M.; Cacan, R.; Verbert, A. Characterization of an oligosaccharide-pyrophosphodolichol pyrophosphatase activity in yeast. Biochem. J. 1988, 255, 235–242. [Google Scholar] [PubMed]
- Massarweh, A.; Bosco, M.; Iatmanen-Harbi, S.; Tessier, C.; Auberger, N.; Busca, P.; Chantret, I.; Gravier-Pelletier, C.; Moore, S.E.H. Demonstration of an oligosaccharide-diphosphodolichol diphosphatase activity whose subcellular localization is different than those of dolichyl-phosphate-dependent enzymes of the dolichol cycle. J. Lipid Res. 2016, 57, 1029–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouhss, A.; Trunkfield, A.E.; Bugg, T.D.H.; Mengin-Lecreulx, D. The biosynthesis of peptidoglycan lipid-linked intermediates. Fems Microbiol. Rev. 2008, 32, 208–233. [Google Scholar] [CrossRef] [PubMed]
- Bosco, M.; Massarweh, A.; Iatmanen-Harbi, S.; Bouhss, A.; Chantret, I.; Busca, P.; Moore, S.E.H.; Gravier-Pelletier, C. Synthesis and biological evaluation of chemical tools for the study of Dolichol Linked Oligosaccharide Diphosphatase (DLODP). Eur. J. Med. Chem. 2017, 125, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Breukink, E.; Wiedemann, I.; van Kraaij, C.; Kuipers, O.P.; Sahl, H.G.; de Kruijff, B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 1999, 286, 2361–2364. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.; Perkins, H.R. Modifications of the acyl-D-alanyl-D-alanine terminus affecting complex-formation with vancomycin. Biochem. J. 1971, 123, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.; Perkins, H.R.; Reynolds, P.E. Reversal by a specific peptide (diacetyl-alpha gamma-L-diaminobutyryl-D-alanyl-D-alanine) of vancomycin inhibition in intact bacteria and cell-free preparations. Biochem. J. 1972, 126, 139–149. [Google Scholar] [CrossRef] [PubMed]
- De Pedro, M.; Schwarz, U. Affinity chromatography of murein precursors on vancomycin-Sepharose. Fems Microbiol. Lett. 1980, 9, 215–217. [Google Scholar] [CrossRef]
- El Ghachi, M.; Bouhss, A.; Barreteau, H.; Touzé, T.; Auger, G.; Blanot, D.; Mengin-Lecreulx, D. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 2006, 281, 22761–22772. [Google Scholar] [CrossRef]
- Alves, G.; Ogurtsov, A.Y.; Yu, Y.-K. Molecular Isotopic Distribution Analysis (MIDAs) with adjustable mass accuracy. J. Am. Soc. Mass Spectrom. 2014, 25, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.T.; Watson, E.E.; Pujari, V.; Conroy, T.; Dowman, L.J.; Giltrap, A.M.; Pang, A.; Wong, W.R.; Linington, R.G.; Mahapatra, S.; et al. Sansanmycin natural product analogues as potent and selective anti-mycobacterials that inhibit lipid I biosynthesis. Nat. Commun. 2017, 8, 14414. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.J.; Gilbey, A.M.; Blewett, A.M.; De Pascale, G.; El Zoeiby, A.; Levesque, R.C.; Catherwood, A.C.; Tomasz, A.; Bugg, T.D.H.; Roper, D.I.; et al. Characterization of tRNA-dependent peptide bond formation by MurM in the synthesis of Streptococcus pneumoniae peptidoglycan. J. Biol. Chem. 2008, 283, 6402–6417. [Google Scholar] [CrossRef] [PubMed]
- Bouhss, A.; Crouvoisier, M.; Blanot, D.; Mengin-Lecreulx, D. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J. Biol. Chem. 2004, 279, 29974–29980. [Google Scholar] [CrossRef] [PubMed]
- Bouhss, A.; Al-Dabbagh, B.; Vincent, M.; Odaert, B.; Aumont-Nicaise, M.; Bressolier, P.; Desmadril, M.; Mengin-Lecreulx, D.; Urdaci, M.C.; Gallay, J. Specific interactions of clausin, a new lantibiotic, with lipid precursors of the bacterial cell wall. Biophys. J. 2009, 97, 1390–1397. [Google Scholar] [CrossRef] [PubMed]
- Abo-Ghalia, M.; Michaud, C.; Blanot, D.; van Heijenoort, J. Specificity of the uridine-diphosphate-N-acetylmuramyl-L-alanyl-D-glutamate: Meso-2,6-diaminopimelate synthetase from Escherichia coli. Eur. J. Biochem. 1985, 153, 81–87. [Google Scholar] [CrossRef]
- Maillard, A.P.; Biarrotte-Sorin, S.; Villet, R.; Mesnage, S.; Bouhss, A.; Sougakoff, W.; Mayer, C.; Arthur, M. Structure-based site-directed mutagenesis of the UDP-MurNAc-pentapeptide-binding cavity of the FemX alanyl transferase from Weissella viridescens. J. Bacteriol. 2005, 187, 3833–3838. [Google Scholar] [CrossRef]
Sample Availability: Samples of compounds are available from the authors. |









| Abbreviation | Compound | Abbreviation | Compound |
|---|---|---|---|
| LII(Lys)55 |  | UDPM5 |  |
| LII(DAP)55 |  | M5P |  |
| LII(DAP)35 |  | GM5P |  |
| LI |  | Ac-KdAdA |  |
| MDP |  | dAdA |  |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massarweh, A.; Bosco, M.; Chantret, I.; Léger, T.; Jamal, L.; Roper, D.I.; Dowson, C.G.; Busca, P.; Bouhss, A.; Gravier-Pelletier, C.; et al. Bacterial Lipid II Analogs: Novel In Vitro Substrates for Mammalian Oligosaccharyl Diphosphodolichol Diphosphatase (DLODP) Activities. Molecules 2019, 24, 2135. https://doi.org/10.3390/molecules24112135
Massarweh A, Bosco M, Chantret I, Léger T, Jamal L, Roper DI, Dowson CG, Busca P, Bouhss A, Gravier-Pelletier C, et al. Bacterial Lipid II Analogs: Novel In Vitro Substrates for Mammalian Oligosaccharyl Diphosphodolichol Diphosphatase (DLODP) Activities. Molecules. 2019; 24(11):2135. https://doi.org/10.3390/molecules24112135
Chicago/Turabian StyleMassarweh, Ahmad, Michael Bosco, Isabelle Chantret, Thibaut Léger, Layla Jamal, David I. Roper, Christopher G. Dowson, Patricia Busca, Ahmed Bouhss, Christine Gravier-Pelletier, and et al. 2019. "Bacterial Lipid II Analogs: Novel In Vitro Substrates for Mammalian Oligosaccharyl Diphosphodolichol Diphosphatase (DLODP) Activities" Molecules 24, no. 11: 2135. https://doi.org/10.3390/molecules24112135
APA StyleMassarweh, A., Bosco, M., Chantret, I., Léger, T., Jamal, L., Roper, D. I., Dowson, C. G., Busca, P., Bouhss, A., Gravier-Pelletier, C., & Moore, S. E. H. (2019). Bacterial Lipid II Analogs: Novel In Vitro Substrates for Mammalian Oligosaccharyl Diphosphodolichol Diphosphatase (DLODP) Activities. Molecules, 24(11), 2135. https://doi.org/10.3390/molecules24112135