Whole Slide Imaging for High-Throughput Sensing Antibiotic Resistance at Single-Bacterium Level and Its Application to Rapid Antibiotic Susceptibility Testing
Abstract
:1. Introduction
2. Results and Discussion
2.1. WSI-Based Monitoring Bacterial Growth at Single-Cell Level
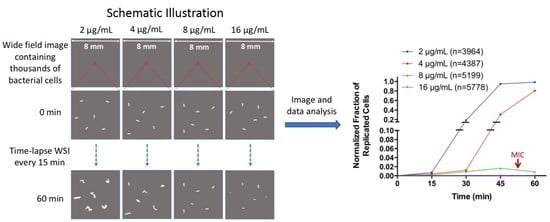
2.2. Determination of Minimal Inhibitory Concentration (MIC) by WSI-Based AST
2.3. Rapid Antibiotic Susceptibility Determination of Phenotypically Heterogeneous Samples
3. Materials and Methods
3.1. Materials and Instruments
3.2. Bacterial Cell Culture
3.3. Sample Preparation
3.4. Whole Slide Imaging
3.5. Image Processing and Data Analysis
3.6. WSI-Based AST
3.7. AST Using Broth Dilution Method
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front Public Health 2014, 2, 145. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Gajdacs, M. The Continuing Threat of Methicillin-Resistant Staphylococcus aureus. Antibiotics 2019, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef] [Green Version]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Lee, N.Y.; Tang, H.J.; Lu, M.C.; Ko, W.C.; Hsueh, P.R. Carbapenem-Resistant Enterobacteriaceae Infections: Taiwan Aspects. Front Microbiol. 2018, 9, 2888. [Google Scholar] [CrossRef] [PubMed]
- Gajdacs, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef]
- Silver, L.L. Challenges of Antibacterial Discovery. Clin. Microbiol. Rev. 2011, 24, 71. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Idelevich, E.A.; Silling, G.; Niederbracht, Y.; Penner, H.; Sauerland, M.C.; Tafelski, S.; Nachtigall, I.; Berdel, W.E.; Peters, G.; Becker, K. Molecular Diagnostics of Sepsis Study, G., Impact of multiplex PCR on antimicrobial treatment in febrile neutropenia: A randomized controlled study. Med. Microbiol. Immunol. 2015, 204, 585–592. [Google Scholar] [CrossRef]
- Kollef, M.H. Inadequate antimicrobial treatment: An important determinant of outcome for hospitalized patients. Clin. Infect. Dis. 2000, 31, S131–S138. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ellis, P.; Arabi, Y.; Roberts, D.; Light, B.; Parrillo, J.E.; Dodek, P.; Wood, G.; Kumar, A.; Simon, D.J.C. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest 2009, 136, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Rolain, J.M.; Mallet, M.N.; Fournier, P.E.; Raoult, D. Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother. 2004, 54, 538–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparbier, K.; Schubert, S.; Weller, U.; Boogen, C.; Kostrzewa, M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J. Clin. Microbiol. 2012, 50, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Vrioni, G.; Tsiamis, C.; Oikonomidis, G.; Theodoridou, K.; Kapsimali, V.; Tsakris, A. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: Current achievements and future perspectives. Ann. Transl. Med. 2018, 6, 240. [Google Scholar] [CrossRef] [PubMed]
- Pulido, M.R.; Garcia-Quintanilla, M.; Martin-Pena, R.; Cisneros, J.M.; McConnell, M.J. Progress on the development of rapid methods for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 2710–2717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoepp, N.G.; Khorosheva, E.M.; Schlappi, T.S.; Curtis, M.S.; Humphries, R.M.; Hindler, J.A.; Ismagilov, R.F. Digital Quantification of DNA Replication and Chromosome Segregation Enables Determination of Antimicrobial Susceptibility after only 15 Minutes of Antibiotic Exposure. Angew. Chem. Int. Ed. Engl. 2016, 55, 9557–9561. [Google Scholar] [CrossRef]
- Schoepp, N.G.; Schlappi, T.S.; Curtis, M.S.; Butkovich, S.S.; Miller, S.; Humphries, R.M.; Ismagilov, R.F. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci. Transl. Med. 2017, 9, 3693. [Google Scholar] [CrossRef]
- Gajdacs, M.; Spengler, G.; Urban, E. Identification and Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Rubik’s Cube of Clinical Microbiology? Antibiotics 2017, 6, 25. [Google Scholar] [CrossRef]
- Hughes, D.; Andersson, D.I. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 374–391. [Google Scholar] [CrossRef] [PubMed]
- Grobner, S.; Dion, M.; Plante, M.; Kempf, V.A. Evaluation of the BD GeneOhm StaphSR assay for detection of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from spiked positive blood culture bottles. J. Clin. Microbiol. 2009, 47, 1689–1694. [Google Scholar] [CrossRef] [PubMed]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Leonard, H.; Colodner, R.; Halachmi, S.; Segal, E. Recent Advances in the Race to Design a Rapid Diagnostic Test for Antimicrobial Resistance. ACS Sens. 2018, 3, 2202–2217. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jung, Y.G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab. Chip. 2013, 13, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jeong, H.Y.; Lee, G.Y.; Han, S.; Han, S.; Jin, B.; Lim, T.; Kim, S.; Kim, D.Y.; Kim, H.C.; et al. Direct, rapid antimicrobial susceptibility test from positive blood cultures based on microscopic imaging analysis. Sci. Rep. 2017, 7, 1148. [Google Scholar] [CrossRef]
- Baltekin, O.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef] [Green Version]
- Cermak, N.; Olcum, S.; Delgado, F.F.; Wasserman, S.C.; Payer, K.R.; Murakami, M.A.; Knudsen, S.M.; Kimmerling, R.J.; Stevens, M.M.; Kikuchi, Y.; et al. High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat. Biotechnol. 2016, 34, 1052–1059. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.; Sarkar, S.; Lin, Z.S.; McKenney, S.; Konry, T. Ultrafast Parallelized Microfluidic Platform for Antimicrobial Susceptibility Testing of Gram Positive and Negative Bacteria. Anal. Chem. 2019, 91, 6242–6249. [Google Scholar] [CrossRef]
- Davies, N.G.; Flasche, S.; Jit, M.; Atkins, K.E. Within-host dynamics shape antibiotic resistance in commensal bacteria. Nat. Ecol. Evol. 2019, 3, 440–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colijn, C.; Cohen, T.; Fraser, C.; Hanage, W.; Goldstein, E.; Givon-Lavi, N.; Dagan, R.; Lipsitch, M. What is the mechanism for persistent coexistence of drug-susceptible and drug-resistant strains of Streptococcus pneumoniae? J. Soc. Interface. 2010, 7, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Sengupta, D.J.; Rosenthal, C.; Costa, G.; Spangler, J.; Sims, E.H.; Jacobs, M.A.; Miller, S.I.; Hoogestraat, D.R.; Cookson, B.T.; et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS ONE 2013, 8, e65226. [Google Scholar] [CrossRef] [PubMed]
- Cummings, L.A.; Kurosawa, K.; Hoogestraat, D.R.; SenGupta, D.J.; Candra, F.; Doyle, M.; Thielges, S.; Land, T.A.; Rosenthal, C.A.; Hoffman, N.G.; et al. Clinical Next Generation Sequencing Outperforms Standard Microbiological Culture for Characterizing Polymicrobial Samples. Clin. Chem. 2016, 62, 1465–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, K.D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006, 70, 660–703. [Google Scholar] [CrossRef] [PubMed]
- Al-Janabi, S.; Huisman, A.; Van Diest, P.J. Digital pathology: Current status and future perspectives. Histopathology 2012, 61, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.; Pantanowitz, L. Current State of the Regulatory Trajectory for Whole Slide Imaging Devices in the USA. J. Pathol. Inform. 2017, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Ghaznavi, F.; Evans, A.; Madabhushi, A.; Feldman, M. Digital imaging in pathology: Whole-slide imaging and beyond. Ann. Rev. Pathol. 2013, 8, 331–359. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Liu, H.; Dong, Q.; Bian, Z.; Wu, H.; Lei, Y. Digital, Rapid, Accurate, and Label-Free Enumeration of Viable Microorganisms Enabled by Custom-Built On-Glass-Slide Culturing Device and Microscopic Scanning. Sensors 2018, 18, 3700. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Wang, Z.; Zhang, Z.; Bian, Z.; Guo, K.; Nambiar, A.; Jiang, Y.; Jiang, S.; Zhong, J.; Choma, M.; et al. Dual light-emitting diode-based multichannel microscopy for whole-slide multiplane, multispectral and phase imaging. J. Biophotonics. 2018, 11, e201700075. [Google Scholar] [CrossRef]
- Liao, J.; Jiang, Y.; Bian, Z.; Mahrou, B.; Nambiar, A.; Magsam, A.W.; Guo, K.; Wang, S.; Cho, Y.K.; Zheng, G. Rapid focus map surveying for whole slide imaging with continuous sample motion. Opt. Lett. 2017, 42, 3379–3382. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Jiang, S.; Zhang, Z.; Guo, K.; Bian, Z.; Jiang, Y.; Zhong, J.; Zheng, G. Terapixel hyperspectral whole-slide imaging via slit-array detection and projection. J. Biomed. Opt. 2018, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, D.; Liu, H.; Ji, H.; Lei, Y. Whole Slide Imaging for High-Throughput Sensing Antibiotic Resistance at Single-Bacterium Level and Its Application to Rapid Antibiotic Susceptibility Testing. Molecules 2019, 24, 2441. https://doi.org/10.3390/molecules24132441
Song D, Liu H, Ji H, Lei Y. Whole Slide Imaging for High-Throughput Sensing Antibiotic Resistance at Single-Bacterium Level and Its Application to Rapid Antibiotic Susceptibility Testing. Molecules. 2019; 24(13):2441. https://doi.org/10.3390/molecules24132441
Chicago/Turabian StyleSong, Donghui, Haomin Liu, Huayi Ji, and Yu Lei. 2019. "Whole Slide Imaging for High-Throughput Sensing Antibiotic Resistance at Single-Bacterium Level and Its Application to Rapid Antibiotic Susceptibility Testing" Molecules 24, no. 13: 2441. https://doi.org/10.3390/molecules24132441
APA StyleSong, D., Liu, H., Ji, H., & Lei, Y. (2019). Whole Slide Imaging for High-Throughput Sensing Antibiotic Resistance at Single-Bacterium Level and Its Application to Rapid Antibiotic Susceptibility Testing. Molecules, 24(13), 2441. https://doi.org/10.3390/molecules24132441