Antibacterial Activity and Molecular Docking Studies of a Selected Series of Hydroxy-3-arylcoumarins
Abstract
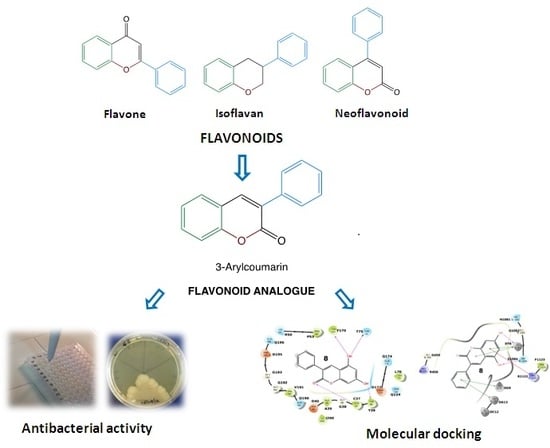
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Chemistry
General Procedure for the Synthesis of Hydroxy-3-arylcoumarins (2–10)
3.2. Biological Studies
3.2.1. Bacterial Cultures
3.2.2. Antioxidant Activity
3.2.3. Statistical Analyses
3.3. Computational Methodology
Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brockhurst, M.A.; Harrison, F.; Veening, J.W.; Harrison, E.; Blackwell, G.; Iqbal, Z.; Maclean, C. Assessing evolutionary risks of resistance for new antimicrobial therapies. Nat. Ecol. Evol. 2019, 3, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Felden, B. Future antibacterial strategies: From basic concepts to clinical challenges. J. Infect. Dis. 2019, 220, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Fair, R.J.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Medicin. Chem. 2014, 6, S14459. [Google Scholar] [CrossRef] [PubMed]
- Simpkin, V.L.; Renwick, M.J.; Kelly, R.; Mossialos, E. Incentivising innovation in antibiotic drug discovery and development: Progress, challenges and next steps. J. Antibiot. (Tokyo) 2017, 70, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, C.; Ardal, C.; Salvesen Blix, H.; Klein, E.; Martinez, E.M.; Lindbaek, M.; Outterson, K.; Rottingen, J.A.; Laxminarayan, R. Introduction and geographic availability of new antibiotics approved between 1999 and 2014. PLoS ONE 2018, 13, e0205166. [Google Scholar] [CrossRef] [PubMed]
- Simoes, N.G.; Bettencourt, A.F.; Monge, N.; Ribeiro, I.A.C. Novel Antibacterial Agents: An Emergent Need to Win the Battle Against Infections. Mini Rev. Med. Chem. 2017, 17, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Garoy, E.Y.; Gebreab, Y.B.; Achila, O.O.; Tekeste, D.G.; Kesete, R.; Ghirmay, R.; Kiflay, R.; Tesfu, T. Methicillin-Resistant Staphylococcus aureus (MRSA): Prevalence and Antimicrobial Sensitivity Pattern among Patients-A Multicenter Study in Asmara, Eritrea. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 8321834. [Google Scholar] [CrossRef]
- Martelli, G.; Giacomini, D. Antibacterial and antioxidant activities for natural and synthetic dual-active compounds. Eur. J. Med. Chem. 2018, 158, 91–105. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, C.; Jang, H.-J.; Kim, B.-o.; Bae, H.-W.; Chung, I.-Y.; Kim, E.S.; Cho, Y.-H. Antibacterial strategies inspired by the oxidative stress and response networks. J. Microbiol. 2019, 57, 203–212. [Google Scholar] [CrossRef]
- Xiao, Z.-P.; Ma, T.-W.; Liao, M.-L.; Feng, Y.-T.; Peng, X.-C.; Li, J.-L.; Li, Z.-P.; Wu, Y.; Luo, Q.; Deng, Y.; et al. Tyrosyl-tRNA synthetase inhibitors as antibacterial agents: Synthesis, molecular docking and structure–activity relationship analysis of 3-aryl-4-arylaminofuran-2(5H)-ones. Eur. J. Med. Chem. 2011, 46, 4904–4914. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Gibson, E.G.; Bax, B.; Chan, P.F.; Osheroff, N. Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase. ACS Infect. Dis. 2019, 5, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.J.; Vazquez-Rodriguez, S.; Fonseca, A.; Uriarte, E.; Santana, L.; Borges, F. Heterocyclic Antioxidants in Nature: Coumarins. Curr. Org. Chem. 2017, 21, 311–324. [Google Scholar] [CrossRef]
- Al-Majedy, Y.K.; Kadhum, A.A.H.; Al-Amiery, A.A.; Mohamad, A.B. Coumarins: The Antimicrobial agents. Syst. Rev. Pharm. 2017, 8, 62–70. [Google Scholar] [CrossRef]
- Jung, J.-W.; Kim, N.-J.; Yun, H.; Han, Y. Recent Advances in Synthesis of 4-Arylcoumarins. Molecules 2018, 23, 2417. [Google Scholar] [CrossRef]
- Veselinović, J.B.; Veselinović, A.M.; Nikolić, G.M.; Pešić, S.Z.; Stojanović, D.B.; Matejić, J.S.; Mihajilov-Krstev, T.M. Antibacterial potential of selected 4-phenyl hydroxycoumarins: Integrated in vitro and molecular docking studies. Med. Chem. Res. 2014, 24, 1626–1634. [Google Scholar] [CrossRef]
- Sun, J.; Ding, W.-X.; Hong, X.-P.; Zhang, K.-Y.; Zou, Y. Synthesis and antimicrobial activities of 4-aryl-3,4-dihydrocoumarins and 4-arylcoumarins. Chem. Nat. Compd. 2012, 48, 16–22. [Google Scholar] [CrossRef]
- Matos, M.J.; Vazquez-Rodriguez, S.; Santana, L.; Uriarte, E.; Fuentes-Edfuf, C.; Santos, Y.; Muñoz-Crego, A. Synthesis and Structure-Activity Relationships of Novel Amino/Nitro Substituted 3-Arylcoumarins as Antibacterial Agents. Molecules 2013, 18, 1394–1404. [Google Scholar] [CrossRef] [Green Version]
- Matos, M.J.; Vazquez-Rodriguez, S.; Santana, L.; Uriarte, E.; Fuentes-Edfuf, C.; Santos, Y.; Munoz-Crego, A. Looking for New Targets: Simple Coumarins as Antibacterial Agents. Med. Chem. 2012, 8, 1140–1145. [Google Scholar] [CrossRef]
- Champlin, F.R.; Ellison, M.L.; Bullard, J.W.; Conrad, R.S. Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 2005, 26, 159–164. [Google Scholar] [CrossRef]
- De Souza, S.M.; Delle Monache, F.; Smania, A., Jr. Antibacterial activity of coumarins. Z. Naturforsch. C 2005, 60, 693–700. [Google Scholar] [CrossRef]
- Gummudavelly, S.; Ranganath, Y.S.; Bhasker, S.; Rajkumar, N. Synthesis and Biological Screening of Some Novel Coumarin Derivatives. Asian J. Res. Chem. 2009, 2, 46–48. [Google Scholar]
- Lin, P.-Y.; Yeh, K.-S.; Su, C.-L.; Sheu, S.-Y.; Chen, T.; Ou, K.-L.; Lin, M.-H.; Lee, L.-W. Synthesis and Antibacterial Activities of Novel 4-Hydroxy-7-hydroxy- and 3-Carboxycoumarin Derivatives. Molecules 2012, 17, 10846–10863. [Google Scholar] [CrossRef] [Green Version]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Matos, M.J.; Varela, C.; Vilar, S.; Hripcsak, G.; Borges, F.; Santana, L.; Uriarte, E.; Fais, A.; Di Petrillo, A.; Pintus, F.; et al. Design and discovery of tyrosinase inhibitors based on a coumarin scaffold. RSC Adv. 2015, 5, 94227–94235. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Robledo-O’Ryan, N.; Matos, M.J.; Vazquez-Rodriguez, S.; Santana, L.; Uriarte, E.; Moncada-Basualto, M.; Mura, F.; Lapier, M.; Maya, J.D.; Olea-Azar, C. Synthesis, antioxidant and antichagasic properties of a selected series of hydroxy-3-arylcoumarins. Biorg. Med. Chem. 2017, 25, 621–632. [Google Scholar] [CrossRef]
- Pintus, F.; Matos, M.J.; Vilar, S.; Hripcsak, G.; Varela, C.; Uriarte, E.; Santana, L.; Borges, F.; Medda, R.; Di Petrillo, A.; et al. New insights into highly potent tyrosinase inhibitors based on 3-heteroarylcoumarins: Anti-melanogenesis and antioxidant activities, and computational molecular modeling studies. Biorg. Med. Chem. 2017, 25, 1687–1695. [Google Scholar] [CrossRef]
- Krishnaswamy, N.R.; Seshadri, T.R.; Sharma, B.R. Study of partial demethylation of some polymethoxy-3-phenylcoumarins and preparation of some new members. Indian J. Chem 1966, 4, 120–126. [Google Scholar]
- Devi, N.; Krishnamurty, H.G. Conversion of 2-methoxychalcones into 3-phenylcoumarins. Indian J. Chem 1994, 33B, 1187–1188. [Google Scholar]
- Pisano, M.B.; Cosentino, S.; Viale, S.; Spanò, D.; Corona, A.; Esposito, F.; Tramontano, E.; Montoro, P.; Tuberoso, C.I.G.; Medda, R.; et al. Biological Activities of Aerial Parts Extracts ofEuphorbia characias. BioMed Res. Int. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Delogu, G.L.; Matos, M.J.; Fanti, M.; Era, B.; Medda, R.; Pieroni, E.; Fais, A.; Kumar, A.; Pintus, F. 2-Phenylbenzofuran derivatives as butyrylcholinesterase inhibitors: Synthesis, biological activity and molecular modeling. Bioorg. Med. Chem. Lett. 2016, 26, 2308–2313. [Google Scholar] [CrossRef]
- Kumar, A.; Pintus, F.; Di Petrillo, A.; Medda, R.; Caria, P.; Matos, M.J.; Vina, D.; Pieroni, E.; Delogu, F.; Era, B.; et al. Novel 2-pheynlbenzofuran derivatives as selective butyrylcholinesterase inhibitors for Alzheimer’s disease. Sci. Rep. 2018, 8, 4424. [Google Scholar] [CrossRef]
- Fais, A.; Kumar, A.; Medda, R.; Pintus, F.; Delogu, F.; Matos, M.J.; Era, B.; Delogu, G.L. Synthesis, molecular docking and cholinesterase inhibitory activity of hydroxylated 2-phenylbenzofuran derivatives. Bioorg. Chem. 2019, 84, 302–308. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Ammarah, U.; Kumar, A.; Pal, R.; Bal, N.C.; Misra, G. Identification of new inhibitors against human Great wall kinase using in silico approaches. Sci. Rep. 2018, 8, 4894. [Google Scholar] [CrossRef]
- Wu, Q.; Peng, Z.; Zhang, Y.; Yang, J. COACH-D: Improved protein-ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018, 46, W438–W442. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Floris, S.; Fais, A.; Rosa, A.; Piras, A.; Marzouki, H.; Medda, R.; González-Paramás, A.M.; Kumar, A.; Santos-Buelga, C.; Era, B. Phytochemical composition and the cholinesterase and xanthine oxidase inhibitory properties of seed extracts from the Washingtonia filifera palm fruit. RSC Adv. 2019, 9, 21278–21287. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds 2–10 are available from the authors. |




| Compounds | A. Gram-Positive Bacterial Strains | |||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus ATCC 25923 | S. aureus TN2A | L. monocytogenes ATCC 19115 | B. cereus ATCC 11178 | |||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| 1 | 500 | >500 | 500 | >500 | 500 | >500 | 500 | >500 |
| 2 | 62.5 | 250 | 62.5 | 250 | 125 | 500 | 15.6 | 62.5 |
| 3 | 62.5 | 500 | 125 | 500 | 250 | >500 | 62.5 | 250 |
| 4 | 125 | 500 | 250 | >500 | 250 | >500 | 62.5 | 250 |
| 5 | 125 | 500 | 125 | 500 | 250 | >500 | 125 | 250 |
| 6 | 500 | >500 | 500 | >500 | 500 | >500 | 250 | 500 |
| 7 | >500 | n.d | >500 | n.d | >500 | n.d | >500 | n.d |
| 8 | 11 | 87.5 | 22 | 87.5 | 44 | 350 | 11 | 44 |
| 9 | >500 | n.d | >500 | n.d | >500 | n.d | >500 | n.d |
| 10 | >500 | n.d | >500 | n.d | >500 | n.d | >500 | n.d |
| Ampicillin a | 2.5 | - | 10 | - | 5 | - | 10 | - |
| Compounds | B. Gram-Negative Bacterial Strains | |||||
|---|---|---|---|---|---|---|
| E. coli ATCC 25922 | S. Enteritidis ATCC 13076 | P. aeruginosa ATCC 27853 | ||||
| MIC | MBC | MIC | MBC | MIC | MBC | |
| 1 | 500 | >500 | 500 | >500 | >500 | n.d |
| 2 | 500 | >500 | >500 | n.d | >500 | n.d |
| 3 | >500 | n.d | >500 | n.d | >500 | n.d |
| 4 | >500 | n.d | >500 | n.d | >500 | n.d |
| 5 | >500 | >500 | >500 | >500 | >500 | n.d |
| 6 | 500 | >500 | 500 | >500 | >500 | n.d |
| 7 | >500 | n.d | >500 | n.d | >500 | n.d |
| 8 | >350 | n.d | >350 | n.d | >350 | n.d |
| 9 | >500 | n.d | >500 | n.d | >500 | n.d |
| 10 | >500 | n.d | >500 | n.d | >500 | n.d |
| Gentamicin a | 5 | - | 5 | - | 5 | - |
| Compounds | EC50 (µM) |
|---|---|
| 2^ | 11.59 ± 0.39 a |
| 3 | 17.42 ± 0.42 b |
| 4 | 23.05 ± 0.43 c |
| 5 | 4.40 ± 0.07 d |
| 8^ | 7.08 ± 0.04 e |
| Trolox $ | 13.0 ± 1.1 a |
| A. Physicochemical Properties | |||
| Compounds | 2 and 8 | 9 | Ampicillin |
| Molecular Formula | C15H10O4 | C15H10O3 | C16H19N3O4S |
| Molecular Weight (g/mol) | 254.24 | 238.24 | 349.40 |
| Rotatable bonds | 1 | 1 | 5 |
| H-bond acceptor atoms | 4 | 3 | 5 |
| H-bond donor atoms | 2 | 1 | 3 |
| Molar refractivity | 71.97 | 69.94 | 92.56 |
| Polar Surface area | 70.67 | 50.44 | 138.03 |
| Lipophilicity (consensus) | 2.46 | 2.91 | 0.08 |
| Water solubility | Soluble | Soluble | Very soluble |
| B. Pharmacokinetics | |||
| Gastrointestinal absorption | High | High | Low |
| Blood–brain barrier permeation | Yes | Yes | No |
| P-glycoprotein substrate | No | No | No |
| Cytochrome P450 1A2 inhibitor | Yes | Yes | No |
| Cytochrome P450 2D6 inhibitor | No | No | No |
| Cytochrome P450 3A4 inhibitor | No | No | No |
| Druglikeness (Lipinski rule) | Yes | Yes | Yes |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pisano, M.B.; Kumar, A.; Medda, R.; Gatto, G.; Pal, R.; Fais, A.; Era, B.; Cosentino, S.; Uriarte, E.; Santana, L.; et al. Antibacterial Activity and Molecular Docking Studies of a Selected Series of Hydroxy-3-arylcoumarins. Molecules 2019, 24, 2815. https://doi.org/10.3390/molecules24152815
Pisano MB, Kumar A, Medda R, Gatto G, Pal R, Fais A, Era B, Cosentino S, Uriarte E, Santana L, et al. Antibacterial Activity and Molecular Docking Studies of a Selected Series of Hydroxy-3-arylcoumarins. Molecules. 2019; 24(15):2815. https://doi.org/10.3390/molecules24152815
Chicago/Turabian StylePisano, Maria Barbara, Amit Kumar, Rosaria Medda, Gianluca Gatto, Rajesh Pal, Antonella Fais, Benedetta Era, Sofia Cosentino, Eugenio Uriarte, Lourdes Santana, and et al. 2019. "Antibacterial Activity and Molecular Docking Studies of a Selected Series of Hydroxy-3-arylcoumarins" Molecules 24, no. 15: 2815. https://doi.org/10.3390/molecules24152815
APA StylePisano, M. B., Kumar, A., Medda, R., Gatto, G., Pal, R., Fais, A., Era, B., Cosentino, S., Uriarte, E., Santana, L., Pintus, F., & Matos, M. J. (2019). Antibacterial Activity and Molecular Docking Studies of a Selected Series of Hydroxy-3-arylcoumarins. Molecules, 24(15), 2815. https://doi.org/10.3390/molecules24152815