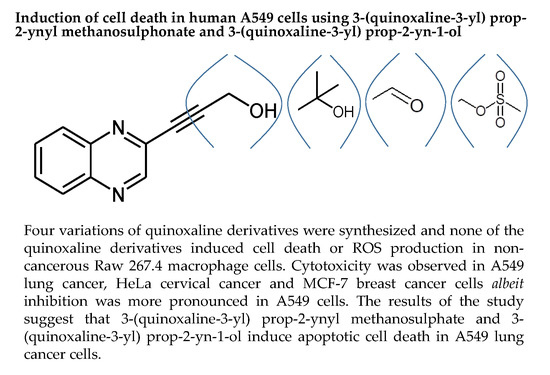
Induction of Cell Death in Human A549 Cells Using 3-(Quinoxaline-3-yl) Prop-2-ynyl Methanosulphonate and 3-(Quinoxaline-3-yl) Prop-2-yn-1-ol
Abstract
:1. Introduction
2. Results
2.1. Chemistry
2.1.1. Procedure for Synthesis of 3-(quinoxalin-3-yl)prop-2-yn-1-ol (LA-55)
2.1.2. Procedure for Synthesis of 2-Methyl-4-(Quinoxalin-3-yl)but-3-yn-2-ol (LA-65C3)
2.1.3. Procedure for Synthesis of 3-(Quinoxalin-3-yl)propionaldehyde (LA-16A)
2.1.4. Procedure for Synthesis of 3-(Quinoxalin-3-yl)Prop-2-Ynyl Methanesulfonate (LA-39B)
2.2. Determination of Reducing Power of Quinoxaline Derivatives
2.3. Determination of Free Radical Scavenging Ability of Quinoxaline Derivatives
2.4. The Effect of Quinoxaline Derivatives LA-39B, LA-55, LA-65C3, and L-16A on Cell Proliferation on HeLa, MCF-7, A549, and Raw 264.7 Cell Lines
2.5. Reduction in ROS Levels in LPS-Stimulated Raw 264.7 Cells Using LA-39B and LA-55
2.6. Reduction in Oxidative Stress on A549 When Treated with LA-39B and LA-55
2.7. Apoptosis Analysis through Nuclear Staining on A549 Cells after Treatment with LA-39B and LA-55
2.8. Apoptosis Analysis through Nuclear Staining on A549 Cells after Treatment with LA-39B and LA-55
3. Discussion
4. Methods
4.1. Ferric Reducing Power Assay (FRAP)
4.2. Quantitative DPPH Assay
4.3. Cell Culture
4.4. MTT Assay
4.5. Analysis of ROS Production
4.6. Apoptosis Analysis using Acridine Orange and Ethidium Bromide
4.7. Annexin V and Dead Cell Apoptosis Analysis
4.8. Designing the Quinoxaline Derivatives
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cervical cancer Incidences, Mortality and Preveence Worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Califano, R.; Landi, L.; Cappuzzo, F. Prognostic and predictive value of K-RAS mutations in non-small cell lung cancer. Drugs 2012, 72, 28–36. [Google Scholar] [CrossRef]
- Shtivelman, E.; Hensing, T.; Simon, G.; Dennis, P.; Otterson, G.; Bueno, R.; Salgia, R. Molecular pathways and therapeutic targets in lung cancer. Oncotargets 2014, 5, 1392–1433. [Google Scholar] [Green Version]
- Subramaniam, S.; Thakur, R.; Yadav, V.; Nanda, R.; Chowdhury, S.; Agrawal, A. Lung cancer biomarkers: State of the art. J. Carcinogen. 2013, 12, 3. [Google Scholar]
- Schiller, J.; Harrington, D.; Belani, C.; Langer, C.; Sandler, A.; Krook, J. Comparison of chemotherapy regimens for advanced non-small cell lung cancer. Nat. Rev. Cancer 2002, 346, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Gargr, V.; Kumar, A.; Chaudhary, A.; Agrawal, S.; Tomar, P.; Sreenivasan, K. Synthesis, biological evaluation and molecular docking studies of 1,3-benzoxazine derivatives as potential anticancer agents. Med. Chem. Res. 2013, 22, 5256–5266. [Google Scholar] [CrossRef]
- Lv, W.; Budke, B.; Pawlowski, M.; Connell, P.P.; Kozikowski, A.P. Development of Small Molecules that Specifically Inhibit the D-loop Activity of RAD51. J. Med. Chem. 2016, 59, 4511–4525. [Google Scholar] [CrossRef] [Green Version]
- Aiello, F.; Carullo, G.; Giordano, F.; Spina, E.; Nigro, A.; Garofalo, A.; Tassini, S.; Constantino, G.; Vincetti, P.; Bruno, A.; et al. Identification of breast cancer inhibitors specific for G protein-coupled estrogen receptor (GPER)-expressing cells. ChemMedChem 2017, 12, 1279–1285. [Google Scholar] [CrossRef]
- Maddila, S.; Momin, M.; Gorle, S.; Palakondu, L.; Jonnalagadda, S. Synthesis and antioxidant evaluation of novel Phenothiazine linked substituted Benzylideneamino-1,2,4-triazole derivatives. J. Chil. Chem. Soc. 2015, 60, 2919–2923. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.C.; Feng, Z.P. The good and bad of microglia/macrophages: New hope in stroke therapeutics. Acta Pharmacol. Sin. 2013, 34, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Nxumalo, W.; Dinsmore, A. Preparation of 6-ethynylpteridine derivatives by Sonogashira coupling. Heterocycles 2013, 87, 79–89. [Google Scholar] [CrossRef]
- Armengol, M.; Joule, J. Synthesis of thieno [2, 3-b] quinoxalines and pyrrolo [1, 2-a] quinoxalines from 2-haloquinoxalines. J. Chem. Soc. Perkin Trans. 2001, 1, 978–984. [Google Scholar] [CrossRef]
- Noolvi, M.; Patel, H.; Bhardwaj, V.; Chauhan, A. Synthesis and in vitro antitumor activity of substituted quinazoline and quinoxaline derivatives: Search for anticancer agent. Eur. J. Med. Chem. 2011, 46, 2327–2346. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashood, S.; Aboldahab, I.; Nagi, M.; Abouzeid, L.; Abdel-Aziz, A.; Abdel-Hamide, S.G.; Youssef, K.; Al-Obaid, A.; El-Subbagh, H. Synthesis, dihydrofolate reductase inhibition, antitumor testing, and molecular modeling study of some new 4(3H)-quinazolinone analogs. Bioorg. Med. Chem. 2006, 14, 8606–8621. [Google Scholar] [CrossRef] [PubMed]
- Vijaya, K.; Sowmyaa, R.-R.; Arathia, B.; Shilpaa, S.; Shwethaa, H.J.; Rajub, M.; Baskaranc, V.; Lakshminarayana, R. Low-dose doxorubicin with carotenoids selectively alters redox status and upregulates oxidative stress-mediated apoptosis in breast cancer cells. Food Chem. Toxicol. 2018, 118, 675–690. [Google Scholar] [CrossRef]
- Moding, E.; Kastan, M.; Kirsch, D. Strategies for optimizing the response of cancer and normal tissues to radiation. Nat. Rev. Drug Discov. 2013, 2, 526–542. [Google Scholar] [CrossRef]
- Benzie, I.; Strain, J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”. The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Blois, M. Antioxidant determinations by the use of a stable free radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Indran, I.R.; Tufo, G.; Pervaiz, S.; Brenner, C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim. Biophys. Acta 2011, 1807, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, F.; Burgeiro, A.; Garcia, R.; Moreno, A.; Carvalho, R.; Oliveira, P. Doxorubicin-induced cardiotoxicity: From bioenergetic failure and cell death ton. Med. Res. Rev. 2014, 34, 106–135. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.; Sánchez-Jiménez, F. Role of reactive oxygen species in apoptosis: Implications for cancer therapy. Int. J. Biochem. Cell Biol. 2000, 32, 157–170. [Google Scholar] [CrossRef]
- Idelchik, M.; Begley, U.; Begley, T.; Melendez, J. Mitochondrial ROS control of cancer. Semin. Cancer Biol. 2017, 47, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Francoa, R.; Sánchez-Oleab, R.; Reyes-Reyesc, E.; Panayiotidis, M. Environmental toxicity, oxidative stress and apoptosis: Ménage à Trois. Mutat. Res. 2009, 674, 3–22. [Google Scholar] [CrossRef]
- Raj, L.; Ide, T.; Gurkar, A.; Foley, M.; Schenone, M.; Li, X. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 2011, 475, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.; Haung, J.; Chang, F.; Huang, K.; Huang, H.; Huang, H. Golden berry-derived 4beta-hydroxywithanolide E for selectively killing oral cancer cells by generating ROS, DNA damage, and apoptotic pathways. PLoS ONE 2013, 8, e64739. [Google Scholar] [CrossRef]
- Suzuki-Karasaki, Y.; Suzuki-Karasaki, M.; Uchida, M.; Ochiai, T. Depolarization controls TRAIL-sensitization and tumor-selective killing of cancer cells: Crosstalk with ROS. Front. Oncol. 2014, 4, 128. [Google Scholar] [CrossRef]
- Baskic, D.; Popovic, S.; Ristic, P.; Arsenijevic, N. Analysis of cycloheximide-induced apoptosis in human leukocytes: Fluorescence microscopy using Annexin V/propidium iodide versus acridin Orange/Ethidium Bromide. Cell Biol. Int. 2006, 30, 924–932. [Google Scholar] [CrossRef]
- Wyllie, A.; Beattie, G.; Hargreaves, A. Chromatin changes in apoptosis. Histochem. J. 1981, 13, 681–692. [Google Scholar] [CrossRef]
- Wong, R. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Srivastava, S.; Banerjee, J.; Srestha, N. Quinoxaline as a potent heterocyclic moiety. J. Pharm. 2014, 4, 17–27. [Google Scholar] [CrossRef]
- Patidar, A.K.; Jeyakandan, M.; Mobiya, A.K.; Selvam, G. Exploring potential of quinoxaline moiety. Int. J. PharmTech Res. 2011, 3, 386–392. [Google Scholar]
- Murthi, P.R.; Suresh, N.; Rani, C.S.; Rao, M.V.B.; Pal, M. Pd/C-catalyzed synthesis of (E)-arylmethylideneidolin-3-ol Under Ultrasound: Their intial evaluation as potential anti-proliferation agents. Lett. Drug Des. Discov. 2015, 12, 109–116. [Google Scholar] [CrossRef]
- Velagapudi, S.; Costales, M.; Vummidi, B.; Nakai, Y.; Angelbello, A.; Tran, T.; Haniff, H.; Matsumoto, Y.; Wang, Z.; Chatterjee, A.; et al. Approved Anti-cancer Drugs Target Oncogenic Non-coding RNAs. Cell Chem. Biol. 2018, 25, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, N.; Heerding, D.A.; Duckett, D.R.; Eberwein, D.J.; Knick, V.B.; Lansing, T.J.; McConnell, R.T.; Gilmer, T.M.; Zhang, S.Y.; Robell, K.; et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 2008, 68, 2366–2374. [Google Scholar] [CrossRef] [PubMed]
- Shareef, H.K.; Muhammed, H.J.; Hussein, H.M.; Hameed, I.H. Antibacterial effect of ginger (Zingiber officinale Roscoe) and bioactive chemical analysis using gas chromatography-mass spectrometry. Orient. J. Chem. 2016, 32, 817–837. [Google Scholar] [CrossRef]
- Lee, M.-Y.; Kim, M.-A.; Kim, H.-J.; Bae, Y.-S.; Kwak, J.-J.; Chung, J.-H.; Yun, J. Alkylation agent Methyl Methanesulfonate (MMS) induces a wave of global protein hyperacetylation: Implication in cancer cell death. Biochem. Biophys. Res. Commun. 2018, 360, 483–489. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |















© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sibiya, M.A.; Raphoko, L.; Mangokoana, D.; Makola, R.; Nxumalo, W.; Matsebatlela, T.M. Induction of Cell Death in Human A549 Cells Using 3-(Quinoxaline-3-yl) Prop-2-ynyl Methanosulphonate and 3-(Quinoxaline-3-yl) Prop-2-yn-1-ol. Molecules 2019, 24, 407. https://doi.org/10.3390/molecules24030407
Sibiya MA, Raphoko L, Mangokoana D, Makola R, Nxumalo W, Matsebatlela TM. Induction of Cell Death in Human A549 Cells Using 3-(Quinoxaline-3-yl) Prop-2-ynyl Methanosulphonate and 3-(Quinoxaline-3-yl) Prop-2-yn-1-ol. Molecules. 2019; 24(3):407. https://doi.org/10.3390/molecules24030407
Chicago/Turabian StyleSibiya, Mixo Aunny, Lerato Raphoko, Dikgale Mangokoana, Raymond Makola, Winston Nxumalo, and Thabe Moses Matsebatlela. 2019. "Induction of Cell Death in Human A549 Cells Using 3-(Quinoxaline-3-yl) Prop-2-ynyl Methanosulphonate and 3-(Quinoxaline-3-yl) Prop-2-yn-1-ol" Molecules 24, no. 3: 407. https://doi.org/10.3390/molecules24030407
APA StyleSibiya, M. A., Raphoko, L., Mangokoana, D., Makola, R., Nxumalo, W., & Matsebatlela, T. M. (2019). Induction of Cell Death in Human A549 Cells Using 3-(Quinoxaline-3-yl) Prop-2-ynyl Methanosulphonate and 3-(Quinoxaline-3-yl) Prop-2-yn-1-ol. Molecules, 24(3), 407. https://doi.org/10.3390/molecules24030407