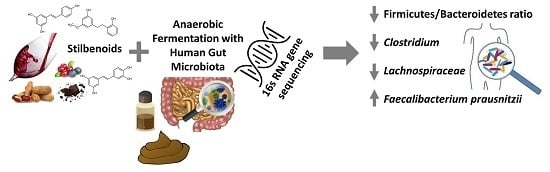
Effect of Selected Stilbenoids on Human Fecal Microbiota
Abstract
:1. Introduction
2. Results and Discussion
2.1. Firmicutes to Bacteroidetes (F/B) Ratio
2.2. Most and Least Abundant Species
2.3. Changes in Relative Abundance (Phylum, Family, Species)
2.3.1. Decrease in Relative Abundance
2.3.2. Increase in Relative Abundance
3. Materials and Methods
3.1. Study Design
3.2. Donors and Ethics Statement
3.3. In vitro Fecal Fermentation (FFM) System
3.3.1. Standard Compounds and Chemicals
3.3.2. Fermentation Medium
3.3.3. Phosphate Buffer, Reducing Solution
3.3.4. Fermentations Using Human Fecal Microbiota
3.4. Microbial Analysis
3.4.1. DNA Extraction
3.4.2. 16.S rDNA amplification: Nested PCR
3.4.3. Semi-conductor Based Next Generation Sequencing
3.4.4. Data Analysis
3.5. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | adolescentis |
| longum | |||||
| Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Eggerthella | lenta | |
| Gen. | sp. | ||||
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | sp. |
| Lactobacillales | Lactobacillaceae | Lactobacillus | sp. | ||
| Leuconostocaceae | Weissella | sp. | |||
| Streptococcaceae | Lactococcus | sp. | |||
| Streptococcus | sp. | ||||
| Clostridia | Clostridiales | [Mogibacteriaceae] | Gen. | sp. | |
| Clostridiaceae | Gen. | sp. | |||
| Lachnospiraceae | [Ruminococcus] | gnavus | |||
| Blautia | sp. | ||||
| sp. | |||||
| Lachnospira | sp. | ||||
| Roseburia | faecis | ||||
| sp. | |||||
| Ruminococcaceae | Ruminococcus | bromii | |||
| sp. | |||||
| Veillonellaceae | Dialister | sp. | |||
| Phascolarctobacterium | sp. | ||||
| Succiniclasticum | sp. | ||||
| Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | [Eubacterium] | biforme | |
| Gen. | sp. | ||||
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | sp. |
| Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Bilophila | sp. | |
| Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Gen. | sp. |
| Phylum | Class | Order | Family | Genus | Species | Stilbenoid | Magnitude Change from 0 h to 24 h | Rel. Abundance at 24 h | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean(%) ± SD | df | Paired-T | Wilcoxon | Paired-T | Wilcoxon | |||||||||
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Control | −20.1 | ± | 9.7 | |||||||
| Res | −22.9 | ± | 10.2 | 3 | 0.025 | 0.068 | 0.045 | 0.068 | ||||||
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Collinsella | aerofaciens | Control | −1.0 | ± | 51.5 | |||||
| Pic | −6.2 | ± | 51.2 | 3 | 0.075 | 0.068 | 0.097 | 0.068 | ||||||
| Gen. | sp. | Control | 27.8 | ± | 80.6 | |||||||||
| Pic | −3.7 | ± | 90.6 | 3 | 0.047 | 0.068 | 0.020 | 0.068 | ||||||
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium | sp. | Control | −54.2 | ± | 28.8 | |||||
| Pic | −62.9 | ± | 28.0 | 3 | 0.029 | 0.068 | 0.098 | 0.068 | ||||||
| Thu | −79.3 | ± | 22.6 | 3 | 0.030 | 0.068 | 0.043 | 0.068 | ||||||
| Lachnospiraceae | [Ruminococcus] | sp. | Control | 32.2 | ± | 68.5 | ||||||||
| Bat | −3.2 | ± | 69.1 | 2 | 0.047 | 0.109 | 0.004 | 0.109 | ||||||
| Pic | −7.0 | ± | 69.4 | 3 | 0.004 | 0.068 | 0.021 | 0.068 | ||||||
| sp. | Control | 15.5 | ± | 20.8 | ||||||||||
| Res | −3.3 | ± | 12.7 | 3 | 0.029 | 0.068 | 0.110 | 0.068 | ||||||
| gnavus | Control | −12.9 | ± | 30.7 | ||||||||||
| Thu | 8.2 | ± | 40.6 | 3 | 0.111 | 0.068 | 0.057 | 0.068 | ||||||
| Blautia | obeum | Control | −29.8 | ± | 35.6 | |||||||||
| Thu | −5.6 | ± | 32.1 | 3 | 0.033 | 0.068 | 0.061 | 0.068 | ||||||
| Coprococcus | sp. | Control | −5.3 | ± | 11.9 | |||||||||
| Res | −19.9 | ± | 3.6 | 3 | 0.063 | 0.068 | 0.030 | 0.068 | ||||||
| Gen. | sp. | Control | −19.0 | ± | 18.4 | |||||||||
| Res | −25.5 | ± | 16.7 | 3 | 0.041 | 0.068 | 0.040 | 0.068 | ||||||
| Faecalibacterium | prausnitzii | Control | −0.5 | ± | 62.5 | |||||||||
| Res | 36.6 | ± | 88.0 | 3 | 0.068 | 0.068 | 0.062 | 0.068 | ||||||
| Paired-T | Wilcoxon | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Stilbenoid | Mean(%) ± SD | t | df | P < 0.05 | P < 0.075 | ||||
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | - | Control | 0.16 | ± | 0.28 | |||||
| Pic | 0.11 | ± | 0.20 | 1.352 | 3 | 0.269 | 0.068 | ||||||
| Coriobacteriia | Coriobacteriales | Coriobacteriaceae | - | Control | 3.68 | ± | 0.93 | ||||||
| Pic | 3.48 | ± | 1.12 | 2.114 | 3 | 0.125 | 0.068 | ||||||
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | - | Control | 0.04 | ± | 0.06 | |||||
| Res | 0.02 | ± | 0.04 | 1.417 | 3 | 0.252 | 0.068 | ||||||
| Leuconostocaceae | - | Control | 0.11 | ± | 0.19 | ||||||||
| Res | 0.13 | ± | 0.21 | −1.733 | 3 | 0.182 | 0.068 | ||||||
| Clostridia | Clostridiales | Lachnospiraceae | - | Control | 43.72 | ± | 4.93 | ||||||
| Res | 42.15 | ± | 4.53 | 3.312 | 3 | 0.045 | 0.068 | ||||||
| Ruminococcaceae | - | Control | 23.70 | ± | 7.06 | ||||||||
| Pic | 27.30 | ± | 2.77 | −1.606 | 3 | 0.207 | 0.068 | ||||||
| Res | 25.39 | ± | 5.64 | −2.062 | 3 | 0.131 | 0.068 | ||||||
| Unnamed | Unnamed | Unnamed | - | Control | 0.01 | ± | 0.00 | ||||||
| Pic | 0.00 | ± | 0.00 | 4.303 | 3 | 0.023 | 0.068 | ||||||
| Phylum | Class | Order | Family | Genus | Species | Stilbenoid | Mean(%) ± SD | df | P < 0.05 | P < 0.075 | |||
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Gen. | sp. | Control | 0.42 | ± | 0.33 | ||||
| Pic | 0.48 | ± | 0.39 | −1.746 | 3 | 0.179 | 0.068 | ||||||
| Res | 0.48 | ± | 0.39 | −1.821 | 3 | 0.166 | 0.068 | ||||||
| Adlercreutzia | sp. | Control | 0.13 | ± | 0.09 | ||||||||
| Pic | 0.09 | ± | 0.07 | 1.554 | 3 | 0.218 | 0.068 | ||||||
| Collinsella | sp. | Control | 0.14 | ± | 0.07 | ||||||||
| Thu | 0.09 | ± | 0.03 | 1.194 | 3 | 0.148 | 0.068 | ||||||
| aerofaciens | Control | 2.58 | ± | 0.55 | |||||||||
| Pic | 2.43 | ± | 0.66 | 2.391 | 3 | 0.097 | 0.068 | ||||||
| Gen. | sp. | Control | 0.13 | ± | 0.05 | ||||||||
| Pic | 0.09 | ± | 0.05 | 4.546 | 3 | 0.020 | 0.068 | ||||||
| Thu | 0.07 | ± | 0.05 | 2.061 | 3 | 0.131 | 0.068 | ||||||
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | sp. | Control | 0.06 | ± | 0.06 | ||||
| Bat | 0.10 | ± | 0.07 | −26.712 | 2 | 0.001 | 0.109 | ||||||
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | Gen. | sp. | Control | 0.03 | ± | 0.06 | ||||
| Res | 0.02 | ± | 0.03 | 1.430 | 3 | 0.248 | 0.068 | ||||||
| Clostridia | Clostridiales | [Mogibacteriaceae] | Gen. | sp. | Control | 0.40 | ± | 0.30 | |||||
| Res | 0.49 | ± | 0.35 | −2.088 | 3 | 0.128 | 0.068 | ||||||
| Clostridiaceae | Clostridium | sp. | Control | 0.07 | ± | 0.05 | |||||||
| Pic | 0.05 | ± | 0.05 | 2.378 | 3 | 0.098 | 0.068 | ||||||
| Res | 0.01 | ± | 0.01 | 1.808 | 3 | 0.168 | 0.068 | ||||||
| Thu | 0.04 | ± | 0.04 | 3.390 | 3 | 0.043 | 0.068 | ||||||
| SMB53 | sp. | Control | 0.04 | ± | 0.01 | ||||||||
| Pic | 0.02 | ± | 0.02 | 2.222 | 3 | 0.113 | 0.068 | ||||||
| Thu | 0.02 | ± | 0.02 | 2.008 | 3 | 0.138 | 0.068 | ||||||
| Gen. | sp. | Control | 0.28 | ± | 0.05 | ||||||||
| Pic | 0.19 | ± | 0.08 | 2.926 | 3 | 0.061 | 0.068 | ||||||
| Lachnospiraceae | [Ruminococcus] | sp. | Control | 0.12 | ± | 0.04 | |||||||
| Bat | 0.09 | ± | 0.05 | 16.420 | 2 | 0.004 | 0.109 | ||||||
| Pic | 0.08 | ± | 0.05 | 4.482 | 3 | 0.021 | 0.068 | ||||||
| Thu | 0.06 | ± | 0.04 | 2.193 | 3 | 0.116 | 0.068 | ||||||
| sp. | Control | 2.17 | ± | 1.71 | |||||||||
| Res | 1.80 | ± | 1.39 | 2.251 | 3 | 0.110 | 0.068 | ||||||
| gnavus | Control | 0.76 | ± | 0.34 | |||||||||
| Thu | 0.91 | ± | 0.33 | -3.012 | 3 | 0.057 | 0.068 | ||||||
| Anaerostipes | sp. | Control | 0.05 | ± | 0.07 | ||||||||
| Pic | 0.04 | ± | 0.07 | 2.485 | 3 | 0.089 | 0.068 | ||||||
| Res | 0.09 | ± | 0.09 | −2.516 | 3 | 0.086 | 0.068 | ||||||
| Blautia | obeum | Control | 0.43 | ± | 0.52 | ||||||||
| Thu | 0.55 | ± | 0.59 | −2.929 | 3 | 0.061 | 0.068 | ||||||
| producta | Control | 0.01 | ± | 0.01 | |||||||||
| Res | 0.00 | ± | 0.00 | 2.185 | 3 | 0.117 | 0.068 | ||||||
| Coprococcus | sp. | Control | 1.88 | ± | 0.54 | ||||||||
| Res | 1.62 | ± | 0.52 | 3.895 | 3 | 0.030 | 0.068 | ||||||
| Dorea | sp. | Control | 0.07 | ± | 0.04 | ||||||||
| Res | 0.07 | ± | 0.04 | -4.817 | 2 | 0.040 | 0.715 | ||||||
| formicigenerans | Control | 0.36 | ± | 0.16 | |||||||||
| Thu | 0.49 | ± | 0.28 | −2.143 | 3 | 0.121 | 0.068 | ||||||
| Lachnospira | sp. | Control | 0.29 | ± | 0.44 | ||||||||
| Pic | 0.33 | ± | 0.48 | −1.723 | 3 | 0.183 | 0.068 | ||||||
| Roseburia | sp. | Control | 0.19 | ± | 0.11 | ||||||||
| Pic | 0.23 | ± | 0.09 | −1.555 | 3 | 0.218 | 0.068 | ||||||
| Gen. | sp. | Control | 12.58 | ± | 0.44 | ||||||||
| Res | 11.59 | ± | 0.80 | 3.468 | 3 | 0.040 | 0.068 | ||||||
| Ruminococcaceae | Faecalibacterium | prausnitzii | Control | 2.01 | ± | 1.01 | |||||||
| Res | 2.79 | ± | 1.53 | −2.912 | 3 | 0.062 | 0.068 | ||||||
| [Mogibacteriaceae] | Gen. | sp. | Control | 0.02 | ± | 0.03 | |||||||
| Res | 0.49 | ± | 0.35 | −2.088 | 3 | 0.128 | 0.068 | ||||||
| Thu | 0.01 | ± | 0.02 | 1.431 | 3 | 0.248 | 0.068 | ||||||
| Unnamed | Unnamed | Unnamed | Gen. | sp. | Control | 0.01 | ± | 0.00 | |||||
| Pic | 0.00 | ± | 0.00 | 4.303 | 3 | 0.023 | 0.068 | ||||||
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Gen. | sp. | Control | 0.01 | ± | 0.01 | ||||
| Thu | 0.00 | ± | 0.00 | 1.884 | 3 | 0.156 | 0.068 | ||||||
| Paired-T | Wilcoxon | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Stilbenoid | Mean(%) ± SD | t | df | P < 0.05 | P < 0.075 | ||||
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | - | Control | 13.01 | ± | 51.37 | |||||
| Pic | 6.73 | ± | 52.38 | 2.465 | 3 | 0.090 | 0.068 | ||||||
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | - | Control | −20.13 | ± | 9.71 | |||||
| Res | −22.88 | ± | 10.24 | 4.197 | 3 | 0.025 | 0.068 | ||||||
| Ruminococcaceae | - | Control | 4.68 | ± | 35.06 | ||||||||
| Pic | 21.94 | ± | 27.47 | −1.604 | 3 | 0.207 | 0.068 | ||||||
| Res | 12.17 | ± | 28.65 | −1.894 | 3 | 0.155 | 0.068 | ||||||
| Phylum | Class | Order | Family | Genus | Species | Stilbenoid | Mean(%) ± SD | df | P < 0.05 | P < 0.075 | |||
| Actinobacteria | Coriobacteria | Coriobacteriales | Coriobacteriaceae | Gen. | sp. | Control | 43.71 | ± | 85.20 | ||||
| Pic | 59.03 | ± | 95.62 | -2.563 | 3 | 0.083 | 0.068 | ||||||
| Res | 58.79 | ± | 92.21 | -2.608 | 3 | 0.080 | 0.068 | ||||||
| Collinsella | Sp. | Control | 24.32 | ± | 63.97 | ||||||||
| Thu | −21.06 | ± | 13.23 | 1.722 | 3 | 0.183 | 0.068 | ||||||
| aerofaciens | Control | −1.03 | ± | 51.47 | |||||||||
| Pic | −6.22 | ± | 51.19 | 2.685 | 3 | 0.075 | 0.068 | ||||||
| Gen. | sp. | Control | 27.75 | ± | 80.59 | ||||||||
| Oxy | −0.93 | ± | 94.16 | 6.272 | 2 | 0.024 | 0.109 | ||||||
| Pic | −3.70 | ± | 90.64 | 3.261 | 3 | 0.047 | 0.068 | ||||||
| Thu | −39.16 | ± | 10.03 | 1.726 | 3 | 0.183 | 0.068 | ||||||
| Firmicutes | Clostridia | Clostridiales | [Mogibacteriaceae] | Gen. | sp. | Control | 72.05 | ± | 96.46 | ||||
| Res | 121.96 | ± | 121.92 | −2.783 | 3 | 0.069 | 0.068 | ||||||
| Clostridiaceae | Clostridium | sp. | Control | −54.19 | ± | 28.78 | |||||||
| Pic | −62.90 | ± | 27.96 | 3.960 | 3 | 0.029 | 0.068 | ||||||
| Res | −90.28 | ± | 15.89 | 1.908 | 3 | 0.152 | 0.068 | ||||||
| Thu | −79.31 | ± | 22.65 | 3.901 | 3 | 0.030 | 0.068 | ||||||
| Gen. | sp. | Control | 122.65 | ± | 206.83 | ||||||||
| Pic | 6.93 | ± | 40.25 | 1.353 | 3 | 0.269 | 0.068 | ||||||
| Lachnospiraceae | [Ruminococcus] | sp. | Control | 32.18 | ± | 68.47 | |||||||
| Bat | −3.23 | ± | 69.11 | 4.448 | 2 | 0.047 | 0.109 | ||||||
| Pic | −7.02 | ± | 69.37 | 8.253 | 3 | 0.004 | 0.068 | ||||||
| Thu | −41.13 | ± | 50.91 | 1.953 | 3 | 0.146 | 0.068 | ||||||
| sp. | Control | 15.46 | ± | 20.76 | |||||||||
| Res | −3.29 | ± | 12.72 | 3.947 | 3 | 0.029 | 0.068 | ||||||
| gnavus | Control | −12.89 | ± | 30.72 | |||||||||
| Thu | 8.24 | ± | 40.57 | −2.244 | 3 | 0.111 | 0.068 | ||||||
| Blautia | obeum | Control | −29.83 | ± | 35.61 | ||||||||
| Thu | −5.56 | ± | 32.11 | −3.763 | 3 | 0.033 | 0.068 | ||||||
| Coprococcus | sp. | Control | −5.31 | ± | 11.92 | ||||||||
| Res | −19.86 | ± | 3.60 | 2.883 | 3 | 0.063 | 0.068 | ||||||
| Dorea | sp. | Control | 16.18 | ± | 75.27 | ||||||||
| Oxy | 59.34 | ± | 95.43 | −9.591 | 2 | 0.011 | 0.109 | ||||||
| formicigenerans | Control | −5.41 | ± | 27.27 | |||||||||
| Thu | 30.22 | ± | 52.23 | −2.397 | 3 | 0.096 | 0.068 | ||||||
| Lachnospira | sp. | Control | 69.70 | ± | 26.12 | ||||||||
| Pic | 128.21 | ± | 95.39 | −1.274 | 3 | 0.292 | 0.068 | ||||||
| Roseburia | sp. | Control | −63.94 | ± | 38.85 | ||||||||
| Pic | −56.62 | ± | 37.62 | −1.597 | 3 | 0.209 | 0.068 | ||||||
| Gen. | sp. | Control | −19.04 | ± | 18.36 | ||||||||
| Res | −25.50 | ± | 16.67 | 3.433 | 3 | 0.041 | 0.068 | ||||||
| Ruminococcaceae | Faecalibacterium | prausnitzii | Control | −0.51 | ± | 62.49 | |||||||
| Res | 36.58 | ± | 87.95 | −2.806 | 3 | 0.068 | 0.068 | ||||||


References
- Edwards, C.A.; Havlik, J.; Cong, W.; Mullen, W.; Preston, T.; Morrison, D.J.; Combet, E. Polyphenols and health: Interactions between fibre, plant polyphenols and the gut microbiota. Nutr. Bull. 2017, 42, 356–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozdal, T.; Sela, D.A.; Xiao, J.; Boyacioglu, D.; Chen, F.; Capanoglu, E. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 2016, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, J.; Fezeu, L.; Touvier, M.; Arnault, N.; Manach, C.; Hercberg, S.; Galan, P.; Scalbert, A. Dietary intake of 337 polyphenols in French adults. Am. J. Clin. Nutr. 2011, 93, 1220–1228. [Google Scholar] [CrossRef] [Green Version]
- Akinwumi, B.C.; Bordun, K.A.M.; Anderson, H.D. Biological activities of stilbenoids. Int. J. Mol. Sci. 2018, 19, 792. [Google Scholar] [CrossRef] [PubMed]
- The WHO Monica Project. A Worlwide Monitoring System for Cardiovascular Diseases: Cardiovascular Mortality & Risk Factors in Selected Communities. In World Health Statistics Annual; WHO: Geneva, Switzerland, 1989; pp. 27–149. [Google Scholar]
- Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C. Interactions of gut microbiota with dietary polyphenols and consequences to human health. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 471–476. [Google Scholar] [CrossRef]
- Carrera-Quintanar, L.; López Roa, R.I.; Quintero-Fabián, S.; Sánchez-Sánchez, M.A.; Vizmanos, B.; Ortuño-Sahagún, D. Phytochemicals That Influence Gut Microbiota as Prophylactics and for the Treatment of Obesity and Inflammatory Diseases. Mediators Inflamm. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.M.L.M.; Bunzel, D.; Huch, M.; Cho, G.G.S.; Ruhland, D.; Bunzel, M.; Bub, A.; Franz, C.M.C.M.A.P.; Kulling, S.E.S.E. In vivo and in vitro metabolism of trans -resveratrol by human gut. Am. J. Clin. Nutr. 2013, 97, 295–309. [Google Scholar] [CrossRef]
- Cueva, C.; Sánchez-Patán, F.; Monagas, M.; Walton, G.E.; Gibson, G.R.; Martín-Álvarez, P.J.; Bartolomé, B.; Moreno-Arribas, M.V. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: Changes in microbial groups and phenolic metabolites. FEMS Microbiol. Ecol. 2013, 83, 792–805. [Google Scholar] [CrossRef]
- Monagas, M.; Urpi-Sarda, M.; Sánchez-Patán, F.; Llorach, R.; Garrido, I.; Gómez-Cordovés, C.; Andres-Lacueva, C.; Bartolomé, B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. [Google Scholar] [CrossRef]
- Guo, W.; Polich, E.D.; Su, J.; Gao, Y.; Christopher, D.M.; Allan, A.M.; Wang, F.; Wang, G.; Zhao, X. The Role of the Gut Microbiota in the Metabolism of Polyphenols as Characterized by Gnotobiotic Mice. Cell Rep. 2015, 11, 1651–1666. [Google Scholar] [CrossRef]
- Etxeberria, U.; Fernández-Quintela, A.; Milagro, F.I.; Aguirre, L.; Martínez, J.A.; Portillo, M.P. Impact of polyphenols and polyphenol-rich dietary sources on gut microbiota composition. J. Agric. Food Chem. 2013, 61, 9517–9533. [Google Scholar] [CrossRef] [PubMed]
- van Duynhoven, J.; Vaughan, E.E.; Jacobs, D.M.; Kemperman, R.A.; van Velzen, E.J.J.; Gross, G.; Roger, L.C.; Possemiers, S.; Smilde, A.K.; Dore, J.; et al. Metabolic fate of polyphenols in the human superorganism. Proc. Natl. Acad. Sci. USA 2011, 108, 4531–4538. [Google Scholar] [CrossRef] [PubMed]
- Espín, J.C.; González-Sarrías, A.; Tomás-Barberán, F.A. The gut microbiota: A key factor in the therapeutic effects of (poly)phenols. Biochem. Pharmacol. 2017, 139, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Cueva, C.; Gil-Sánchez, I.; Ayuda-Durán, B.; González-Manzano, S.; González-Paramás, A.M.; Santos-Buelga, C.; Bartolomé, B.; Victoria Moreno-Arribas, M. An integrated view of the effects of wine polyphenols and their relevant metabolites on gut and host health. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, M.; Muñoz-González, I.; Cueva, C.; Jiménez-Girón, A.; Sánchez-Patán, F.; Santos-Buelga, C.; Moreno-Arribas, M.V.; Bartolomé, B. A survey of modulation of gut microbiota by dietary polyphenols. Biomed. Res. Int. 2015, 8050902. [Google Scholar] [CrossRef] [PubMed]
- Larrosa, M.; Yañéz-Gascón, M.J.; Selma, M.V.; González-Sarrías, A.; Toti, S.; Cerón, J.J.; Tomás-Barberán, F.; Dolara, P.; Espín, J.C. Effect of a low dose of dietary resveratrol on colon microbiota, inflammation and tissue damage in a DSS-induced colitis rat model. J. Agric. Food Chem. 2009, 57, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Jiang, H.; Fang, J. Regulation of Immune Function by Polyphenols. J. Immunol. Res. 2018, 2018, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braune, A.; Blaut, M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 2016, 7, 216–234. [Google Scholar] [CrossRef] [Green Version]
- Tomas-Barberan, F.A.; Selma, M.V.; Espín, J.C. Polyphenols’ Gut Microbiota Metabolites: Bioactives or Biomarkers? J. Agric. Food Chem. 2018, 66, 3593–3594. [Google Scholar] [CrossRef]
- Marín, L.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Bioavailability of dietary polyphenols and gut microbiota metabolism: Antimicrobial properties. Biomed Res. Int. 2015. [Google Scholar] [CrossRef]
- Kemperman, R.A.; Gross, G.; Mondot, S.; Possemiers, S.; Marzorati, M.; Van de Wiele, T.; Doré, J.; Vaughan, E.E. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res. Int. 2013, 53, 659–669. [Google Scholar] [CrossRef]
- Volstatova, T.; Marsik, P.; Rada, V.; Geigerova, M.; Havlik, J. Effect of apple extracts and selective polyphenols on the adhesion of potential probiotic strains of Lactobacillus gasseri R and Lactobacillus casei FMP. J. Funct. Foods 2017, 35, 391–397. [Google Scholar] [CrossRef]
- Havlik, J.; Edwards, C.A. Non-extractable Polyphenols into Polyphenol Research. In Non-extractable Polyphenols and Carotenoids; RSC Publishing: Cambridge, UK, 2018; pp. 241–262. ISBN 9781788013208. [Google Scholar]
- Tzounis, X.; Vulevic, J.; Kuhnle, G.G.C.; George, T.; Leonczak, J.; Gibson, G.R.; Kwik-Uribe, C.; Spencer, J.P.E. Flavanol monomer-induced changes to the human faecal microflora. Br. J. Nutr. 2008, 99, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Mayta-Apaza, A.C.; Pottgen, E.; De Bodt, J.; Papp, N.; Marasini, D.; Howard, L.; Abranko, L.; Van de Wiele, T.; Lee, S.O.; Carbonero, F. Impact of tart cherries polyphenols on the human gut microbiota and phenolic metabolites in vitro and in vivo. J. Nutr. Biochem. 2018, 59, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Catinean, A.; Neag, M.A.; Muntean, D.M.; Bocsan, I.C.; Buzoianu, A.D. An overview on the interplay between nutraceuticals and gut microbiota. PeerJ 2018, 6, e4465. [Google Scholar] [CrossRef] [PubMed]
- Bialonska, D.; Kasimsetty, S.G.; Schrader, K.K.; Ferreira, D. The effect of pomegranate (punica granatum l.) byproducts and ellagitannins on the growth of human gut bacteria. J. Agric. Food Chem. 2009, 57, 8344–8349. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, C.; Marzorati, M.; Innocenti, M.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Van De Wiele, T.; Mulinacci, N. Dietary supplement based on stilbenes: A focus on gut microbial metabolism by the: In vitro simulator M-SHIME®. Food Funct. 2016, 7, 4564–4575. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Scarano, A.; Butelli, E.; De Santis, S.; Cavalcanti, E.; Hill, L.; De Angelis, M.; Giovinazzo, G.; Chieppa, M.; Martin, C.; Santino, A. Combined Dietary Anthocyanins, Flavonols, and Stilbenoids Alleviate Inflammatory Bowel Disease Symptoms in Mice. Front. Nutr. 2018, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hervert-Hernández, D.; Goñi, I. Dietary polyphenols and human gut microbiota: A review. Food Rev. Int. 2011, 27, 154–169. [Google Scholar] [CrossRef]
- Etxeberria, U.; Hijona, E.; Aguirre, L.; Milagro, F.I.; Bujanda, L.; Rimando, A.M.; Martínez, J.A.; Portillo, M.P. Pterostilbene-induced changes in gut microbiota composition in relation to obesity. Mol. Nutr. Food Res. 2017, 61, 1500906. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Healey, G.R.; Murphy, R.; Brough, L.; Butts, C.A.; Coad, J. Interindividual variability in gut microbiota and host response to dietary interventions. Nutr. Rev. 2017, 75, 1059–1080. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 4–9. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027. [Google Scholar] [CrossRef]
- Qiao, Y.; Sun, J.; Xia, S.; Tang, X.; Shi, Y.; Le, G. Effects of resveratrol on gut microbiota and fat storage in a mouse model with high-fat-induced obesity. Food Funct. 2014, 5, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut Microbiota and Extreme Longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2003, 31, 23–27. [Google Scholar] [CrossRef] [Green Version]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012, 40, 5–15. [Google Scholar] [CrossRef]
- Kong, F.; Hua, Y.; Zeng, B.; Ning, R.; Li, Y.; Zhao, J. Gut microbiota signatures of longevity. Curr. Biol. 2016, 26, R832–R833. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, L. Host-microbiome interactions in health and disease. Clin. Liver Dis. 2015, 5, 142–144. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, B.S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 2013, 28, 9–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiyama, S.; Ueno, T.; Suzuki, T. Identification of a newly isolated equol-producing lactic acid bacterium from the human feces. J. Intest. Microbiol. 2007, 21, 217–220. [Google Scholar]
- Gaya, P.; Medina, M.; Sánchez-Jiménez, A.; Landete, J. Phytoestrogen Metabolism by Adult Human Gut Microbiota. Molecules 2016, 21, 1034. [Google Scholar] [CrossRef] [PubMed]
- Summaries, S. Dysbiosis of the Faecal Microbiota in Patients with Crohn’s Disease and Their Unaffected Relatives. Gut 2011, 9, 166–168. [Google Scholar]
- Jeffery, I.B.; Claesson, M.J.; O′toole, P.W. Categorization of the gut microbiota: Enterotypes or gradients? Grouping the microbiota of individual subjects into compositional categories, or enterotypes, based on the dominance of certain genera may have oversimplified a complex situation. Nat. Rev. Microbiol. 2012, 10, 591–592. [Google Scholar] [CrossRef]
- Lawson, P.A.; Finegold, S.M. Reclassification of Ruminococcus obeum as Blautia obeum comb. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 789–793. [Google Scholar] [CrossRef]
- Shortt, C.; Hasselwander, O.; Meynier, A.; Nauta, A.; Fernández, E.N.; Putz, P.; Rowland, I.; Swann, J.; Türk, J.; Vermeiren, J.; et al. Systematic review of the effects of the intestinal microbiota on selected nutrients and non-nutrients. Eur. J. Nutr. 2018, 57, 25–49. [Google Scholar] [CrossRef]
- Al Shukor, N.; Van Camp, J.; Gonzales, G.B.; Staljanssens, D.; Struijs, K.; Zotti, M.J.; Raes, K.; Smagghe, G. Angiotensin-converting enzyme inhibitory effects by plant phenolic compounds: A study of structure activity relationships. J. Agric. Food Chem. 2013, 61, 11832–11839. [Google Scholar] [CrossRef]
- Karamać, M.; Amarowicz, R. Inhibition of Pancreatic Lipase by Phenolic Acids-Examination in vitro. Zeitschrift fur Naturforsch. Sect. C J. Biosci. 1996, 51, 903–906. [Google Scholar] [CrossRef]
- Sandra Goncalves, A.R. Inhibitory Properties of Phenolic Compounds Against Enzymes Linked with Human Diseases. In Phenolic Compounds-Biological Activity; Soto-Hernandez, M., Tenango, M.P., García-Mateos, R., Eds.; InTech: London, UK, 2017; Volume 2, pp. 581–770. ISBN 9789537619992. [Google Scholar]
- McMurry, J. Fundamentals of Organic Chemistry, 4th Ed.; Brooks/Cole Publishing Company: Pacific Grove, CA, USA, 1998; ISBN 0534352154. [Google Scholar]
- Iuga, C.; Alvarez-Idaboy, J.R.; Russo, N. Antioxidant Activity of trans -Resveratrol toward Hydroxyl and Hydroperoxyl Radicals: A Quantum Chemical and Computational Kinetics Study. J. Org. Chem. 2012, 77, 3868–3877. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, T.; Melzig, M.F. Polyphenolic Compounds as Pancreatic Lipase Inhibitors. Planta Med. 2015, 81, 771–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Ni, X.; Kai, G.; Chen, X. A review on structure-activity relationship of dietary polyphenols inhibiting α-amylase. Crit. Rev. Food Sci. Nutr. 2013, 53, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Tadera, K.; Minami, Y.; Takamatsu, K.; Matsuoka, T. Inhibition of α-Glucosidase and α-Amylase by Flavonoids. J. Nutr. Sci. Vitaminol. (Tokyo) 2006, 52, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Lo Piparo, E.; Scheib, H.; Frei, N.; Williamson, G.; Grigorov, M.; Chou, C.J. Flavonoids for controlling starch digestion: Structural requirements for inhibiting human α-amylase. J. Med. Chem. 2008, 51, 3555–3561. [Google Scholar] [CrossRef] [PubMed]
- Burapan, S.; Kim, M.; Han, J. Curcuminoid Demethylation as an Alternative Metabolism by Human Intestinal Microbiota. J. Agric. Food Chem. 2017, 65, 3305–3310. [Google Scholar] [CrossRef] [PubMed]
- Blaut, M.; Schoefer, L.; Braune, A. Transformation of Flavonoids by Intestinal Microorganisms. Int. J. Vitam. Nutr. Res. 2003, 73, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.S.; Zhao, Y.F.; Nakamura, N.; Akao, T.; Kakiuchi, N.; Min, B.S.; Hattori, M. Enantioselective Dehydroxylation of Enterodiol and Enterolactone Precursors by Human Intestinal Bacteria. Biol. Pharm. Bull. 2007, 30, 2113–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.-Q.; Meselhy, M.R.; Li, Y.; Qin, G.-W.; HAattori, M. Human intestinal bacteria capable of transforming secoisolariciresinol diglucoside to mammalian lignans, enterodiol and enterolactone. Chem. Pharm. Bull. 2000, 48, 1606–1610. [Google Scholar] [CrossRef] [PubMed]
- Etxeberria, U.; Arias, N.; Boqué, N.; Macarulla, M.T.; Portillo, M.P.; Martínez, J.A.; Milagro, F.I. Reshaping faecal gut microbiota composition by the intake of trans -resveratrol and quercetin in high-fat sucrose diet-fed rats. J. Nutr. Biochem. 2015, 26, 651–660. [Google Scholar] [CrossRef] [PubMed]
- González-Barrio, R.; Edwards, C.A.; Crozier, A. Colonic catabolism of ellagitannins, ellagic acid, and raspberry anthocyanins: In vivo and in vitro studies. Drug Metab. Dispos. 2011, 39, 1680–1688. [Google Scholar] [CrossRef] [PubMed]
- Jaganath, I.B.; Mullen, W.; Lean, M.E.J.; Edwards, C.A.; Crozier, A. In vitro catabolism of rutin by human fecal bacteria and the antioxidant capacity of its catabolites Indu. Free Radic. Biol. Med. 2009, 47, 1180–1189. [Google Scholar] [CrossRef]
- Edwards, C.A.; Gibson, G.; Champ, M.; Jensen, B.-B.; Mathers, J.C.; Nagengast, F.; Rumney, C.; Quehl, A. In Vitro Method for Quantification of the Fermentation of Starch by Human Faecal Bacteria. J. Sci. Food Agric. 1996, 71, 209–217. [Google Scholar] [CrossRef]
- Juretschko, S.; Timmermann, G.; Schmid, M.; Schleifer, K.; Pommerening-ro, A. Combined Molecular and Conventional Analyses of Nitrifying Bacterium Diversity in Activated Sludge: Nitrosococcus mobilis and Nitrospira-Like Bacteria as Dominant Populations. Appl. Environ. Microbiol. 1998, 64, 3042–3051. [Google Scholar] [PubMed]
- Fliegerova, K.; Tapio, I.; Bonin, A.; Mrazek, J.; Callegari, M.L.; Bani, P.; Bayat, A.; Vilkki, J.; Kopečný, J.; Shingfield, K.J.; et al. Effect of DNA extraction and sample preservation method on rumen bacterial population. Anaerobe 2014, 29, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Milani, C.; Hevia, A.; Foroni, E.; Duranti, S.; Turroni, F.; Lugli, G.A.; Sanchez, B.; Martín, R.; Gueimonde, M.; van Sinderen, D.; et al. Assessing the Fecal Microbiota: An Optimized Ion Torrent 16S rRNA Gene-Based Analysis Protocol. PLoS ONE 2013, 8, e68739. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. correspondence QIIME allows analysis of high- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nature 2010, 7, 335–336. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |




| Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | sp. |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | sp. |
| Clostridia | Clostridiales | Lachnospiraceae | Blautia | sp. | |
| Gen. | sp. | ||||
| Gen. | sp. | ||||
| Ruminococcaceae | Faecalibacterium | prausnitzii | |||
| Ruminococcus | sp. | ||||
| Gen. | sp. | ||||
| Gen. | sp. | ||||
| Unnamed | Gen. | sp. | |||
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Gen. | sp. |
| Stilbenoid | Effect | Phylum | Family | Genus | Species | Notes | |
|---|---|---|---|---|---|---|---|
| Resveratrol | ↑ | Actinobacteria | Bifidobacteriaceae | Bifidobacterium | sp. | NS | |
| Firmicutes | Clostridiaceae | Clostridium | XB90 | S | |||
| Faecalibacterium | prausnitzii | S | Won’t grow without acetate in pure culture. | ||||
| Lactobacillaceae | Lactobacillus | sp. | Un. | ||||
| ↓ | Bacteroidetes | Tannerellaceae | Parabacteroides | distansonis | NS | Only detected in one donor. | |
| Firmicutes | Clostridiaceae | Clostridium | aldenense | S | Species not identified, responsesignificn at genus level. | ||
| C9 | S | ||||||
| hathewayi | S | ||||||
| MLG661 | S | ||||||
| Enterococcaceae | Enterococcus | faecalis | ND | ||||
| Gracilibacteraceae | Gracilibacter | thermotolerans | ND | ||||
| Proteobacteria | Enterobacteriaceae | Proteus | mirabilis | ND | |||
| Firmicutes to Bacteroidetes (F/B) ratio | S | ||||||
| Other | Actinobacteria | Coriobacteriaceae | Slackia | equolifaciens | Other | Dihydroresveratrol producers. Identified at genus level only. Slackia’s abundance highest for Res, and not detectable at Ctrl0. Adlercreutzia’s abundance highest for Ctrl24, and lowest for Ctrl0. | |
| Adlercreutzia | equolifaciens | Other | |||||
| Phenolic mix, includes Resveratrol | ↑ | Verrucomicrobia | Verrucomicrobiaceae | Akkermansia | muciniphila | S | Mice study. Detected in one of our donors. |
| ↓ | Firmicutes | Lachnospiraceae | Blautia | sp. | Un. | Mice study. | |
| Ruminococcaceae | Oscillospira | sp. | S | Mice study. Has never been cultured, but always detected. | |||
| Piceatannol | ↑ | Firmicutes | Lactobacillaceae | Lactobacillus | sp. | NS | Mice study. |
| Unnamed | Gen. | sp. | NS | Mice study. | |||
| ↓ | Bacteroidetes | Unnamed | Gen. | sp. | NS | Mice study. Decrease was observed, but at a lower magnitude than Ctrl24. | |
| Other | Bacteroidaceae | Gen. | sp. | S | Mice study. Abundance change. | ||
| Fiber | ↑ | Bacteroidetes | Prevotellaceae | Prevotella | sp. | S | Stilbenoids associated with fiber-containing food. |
| Plant-based diet | Firmicutes | Clostridiaceae | Faecalibacterium | prausnitzii | S | Saccharolytic microbes. | |
| Lachnospiraceae | Roseburia | sp. | NS | Saccharolytic microbes. | |||
| ↓ | Proteobacteria | Desulfovibrionaceae | Bilophila | sp. | ND | Putrefactive microbes. Less abundance expected in a plant-based diet. | |
| Bacteroidetes | Bacteroidaceae | Bacteroides | sp. | NS | Putrefactive microbes. Less abundance expected in a plant-based diet. | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaimes, J.D.; Jarosova, V.; Vesely, O.; Mekadim, C.; Mrazek, J.; Marsik, P.; Killer, J.; Smejkal, K.; Kloucek, P.; Havlik, J. Effect of Selected Stilbenoids on Human Fecal Microbiota. Molecules 2019, 24, 744. https://doi.org/10.3390/molecules24040744
Jaimes JD, Jarosova V, Vesely O, Mekadim C, Mrazek J, Marsik P, Killer J, Smejkal K, Kloucek P, Havlik J. Effect of Selected Stilbenoids on Human Fecal Microbiota. Molecules. 2019; 24(4):744. https://doi.org/10.3390/molecules24040744
Chicago/Turabian StyleJaimes, Jose D., Veronika Jarosova, Ondrej Vesely, Chahrazed Mekadim, Jakub Mrazek, Petr Marsik, Jiri Killer, Karel Smejkal, Pavel Kloucek, and Jaroslav Havlik. 2019. "Effect of Selected Stilbenoids on Human Fecal Microbiota" Molecules 24, no. 4: 744. https://doi.org/10.3390/molecules24040744
APA StyleJaimes, J. D., Jarosova, V., Vesely, O., Mekadim, C., Mrazek, J., Marsik, P., Killer, J., Smejkal, K., Kloucek, P., & Havlik, J. (2019). Effect of Selected Stilbenoids on Human Fecal Microbiota. Molecules, 24(4), 744. https://doi.org/10.3390/molecules24040744