Enantiomeric Tartaric Acid Production Using cis-Epoxysuccinate Hydrolase: History and Perspectives
Abstract
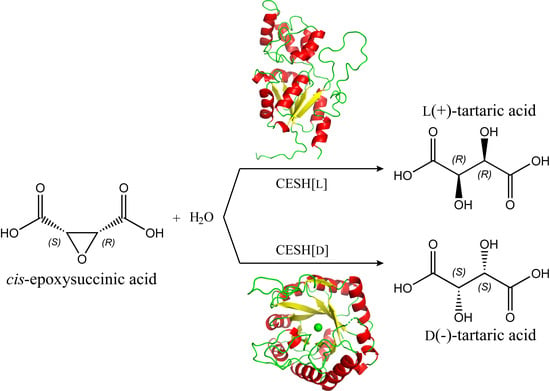
:1. Introduction
2. The History of CESH Studies
3. Bacteria that Produce CESHs
4. Stability of CESHs
5. Process Optimization for TA Production Using CESHs
6. Structure and Catalytic Mechanism of CESH[L]
7. Structure and Catalytic Mechanism of CESH[D]
8. Perspective for CESH Research and Application
Funding
Conflicts of Interest
References
- Derewenda, Z.S. On wine, chirality and crystallography. Acta Crystallogr. Sect. A: Found. Crystallogr. 2008, 64, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Kodama, S.; Yamamoto, A.; Matsunaga, A.; Hayakawa, K. Direct chiral resolution of tartaric acid in food products by ligand exchange capillary electrophoresis using copper(Il)-d-quinic acid as a chiral selector. J. Chromatogr. 2001, 932, 139–143. [Google Scholar] [CrossRef]
- Willaert, R.; De Vuyst, L. Continuous production of l(+)-tartaric acid from cis-epoxysuccinate using a membrane recycle reactor. Appl. Microbiol. Biotechnol. 2006, 71, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Skarzewski, J.; Gupta, A. Synthesis of C2 symmetric primary vicinal diamines. Double stereospecific Mitsunobu reaction on the heterocyclic diols derived from tartaric acid. Tetrahedron Asymmetry 1997, 8, 1861–1867. [Google Scholar] [CrossRef]
- Su, Y.L.; Yang, C.S.; Teng, S.J.; Zhao, G.; Ding, Y. Total synthesis of four diastereoisomers of Goniofufurone from d-(−)- or l-(+)-tartaric acid. Tetrahedron 2001, 57, 2147–2153. [Google Scholar] [CrossRef]
- Pabba, J.; Vasella, A. Synthesis of d-gluco-, l-ido-, d-galacto-, and l-altro-configured glycaro-1,5-lactams from tartaric acid. Tetrahedron Lett. 2005, 46, 3619–3622. [Google Scholar] [CrossRef]
- Gawronski, J.; Gawronska, K. Tartaric and Malic Acids in Synthesis: A Source Book of Building Blocks, Ligands, Auxiliaries, and Resolving Agents, 1st ed.; Wiley-Interscience: New York, NY, USA, 1999. [Google Scholar]
- Ghosh, A.K.; Koltun, E.S.; Bilcer, G. Tartaric acid and tartrates in the synthesis of bioactive molecules. Synthesis-Stuttgart 2001, 2001, 1281–1301. [Google Scholar] [CrossRef] [PubMed]
- Gratzer, K.; Gururaja, G.N.; Waser, M. Towards tartaric-acid-derived asymmetric organocatalysts. Eur. J. Org. Chem. 2013, 2013, 4471–4482. [Google Scholar] [CrossRef] [PubMed]
- Mikova, H.; Rosenberg, M.; Kristofikova, L.; Liptaj, T. Influence of molybdate and tungstate ions on activity of cis-epoxysuccinate hydrolase in Nocardia tartaricans. Biotechnol. Lett. 1998, 20, 833–835. [Google Scholar] [CrossRef]
- Steinreiber, A.; Faber, K. Microbial epoxide hydrolases for preparative biotransformations. Curr. Opin. Biotechnol. 2001, 12, 552–558. [Google Scholar] [CrossRef]
- Blee, E.; Schuber, F. Stereocontrolled hydrolysis of the linoleic-acid monoepoxide regioisomers catalyzed by soybean epoxide hydrolase. Eur. J. Biochem. 1995, 230, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Linderman, R.J.; Walker, E.A.; Haney, C.; Roe, R.M. Determination of the regiochemistry of insect epoxide hydrolase catalyzed epoxide hydration of juvenile-hormone by 18O-labeling studies. Tetrahedron 1995, 51, 10845–10856. [Google Scholar] [CrossRef]
- Weijers, C.A.G.M.; de Bont, J.A.M. Epoxide hydrolases from yeasts and other sources: Versatile tools in biocatalysis. J. Mol. Catal. B Enzym. 1999, 6, 199–214. [Google Scholar] [CrossRef]
- Arand, M.; Cronin, A.; Adamska, M.; Oesch, F. Epoxide hydrolases: Structure, function, mechanism, and assay. Methods Enzymol. 2005, 400, 569–588. [Google Scholar] [PubMed]
- Faber, K.; Mischitz, M.; Kroutil, W. Microbial epoxide hydrolases. Acta Chem. Scand. 1996, 50, 249–258. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Xu, Y.; Ping, L.; Zheng, Y. Cloning, sequencing, and expression of a novel epoxide hydrolase gene from Rhodococcus opacus in Escherichia coli and characterization of enzyme. Appl. Microbiol. Biotechnol. 2007, 74, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, T.; Lu, H.; Ma, X.; Kai, L.; Guo, K.; Zhao, Y. Purification and characterization of a cis-epoxysuccinic acid hydrolase from Bordetella sp. strain 1-3. Protein Expression Purif. 2010, 69, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Bao, W.; Xie, Z.; Zhang, J.; Li, Y. Molecular cloning and characterization of a cis-epoxysuccinate hydrolase from Bordetella sp. BK-52. J. Microbiol. Biotechnol. 2010, 20, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Archelas, A.; Furstoss, R. Epoxide hydrolases: New tools for the synthesis of fine organic chemicals. Trends Biotechnol. 1998, 16, 108–116. [Google Scholar] [CrossRef]
- Sato, E.; Yanai, A. Method for Preparing d-tartaric Acid. U.S. Patent 3,957,579, 18 May 1976. CAN 84:89630. [Google Scholar]
- Kamatani, Y.; Okazaki, H.; Imai, K.; Fujita, N.; Yamazaki, Y.; Ogino, K. Production of l(+)-tartaric Acid. U.S. Patent 4,011,135, 8 March 1977. CAN 85:175611. [Google Scholar]
- Kamatani, Y.; Okazaki, H.; Imai, K.; Fujita, N.; Yamazaki, Y.; Ogino, K. Method for producing l(+)-tartaric acid. U.S. Patent 4,028,185, 7 June 1977. CAN 85:175611. [Google Scholar]
- Miura, Y.; Yutani, K.; Izumi, Y. Process for preparing L-tartaric acid. U.S. Patent 4,010,072, 1 March 1977. CAN 86:15234. [Google Scholar]
- Miura, Y.; Yutani, K.; Takesue, H.; Fujii, K. Microbiological process for preparing l-tartaric acid in presence of surfactants. U.S. Patent 4,017,362, 12 April 1977. [Google Scholar]
- Tsurumi, Y.; Fujioka, T. Process for manufacture of l(+)-tartaric acid or salts thereof. U.S. Patent 4,092,220, 30 May 1978. [Google Scholar]
- Huang, T.; Qian, X. Production of l(+)-tartaric acid. Ind. Microbiol. 1990, 20, 14–17. [Google Scholar]
- Zhang, J.; Huang, T. Productivity of l(+)-tartaric acid using microbial conversion method. Ind. Microbiol. 1990, 20, 7–12, 24. [Google Scholar]
- Zheng, P.; Sun, Z. Enzymatic conversion of cis-epoxysuccinate into l(+)-tartarate by Nocardia sp. SW13-57. Ind. Microbiol. 1994, 24, 12–17. [Google Scholar]
- Sun, Z.; Zheng, P.; Dai, X.; Li, H.; Jin, M. Production of l(+)-tartaric acid by immobilized Nocardia tartaricans SW13-57. Chin. J. Biotechnol./Shengwu Gongcheng Xuebao 1995, 11, 372–376. [Google Scholar]
- Yamagishi, K.; Cho, H.; Takai, Y.; Kawaguchi, T. Production of d(−)-tartaric acid. Jp. Patent 08-245497, 24 September 1996. CAN 126:7698. [Google Scholar]
- Lou, J.-F.; Zhang, J.-G. Research on microbial productions of l(+)-tartaric acid. Food Sci. Technol. 2006, 2006, 162–164. [Google Scholar]
- Ikuta, M.; Sakamoto, T.; Ueda, M.; Sashita, R. Production of d(−)-tartaric acid. Jp. Patent 2000-014391, 18 January 2000. CAN 132:92374. [Google Scholar]
- Pan, K.-X.; Min, H.; Xia, Y.; Xu, X.-Y.; Ruan, A.-D. Isolation, identification and phylogenetic analysis of Rhodococcus sp. strain M1 producing cis-epoxysuccinate hydrolase and optimization of production conditions. Acta Microbiol. Sin. 2004, 44, 276–280. [Google Scholar]
- Li, X.; Ma, X.; Zhao, Y.; Jia, X.; Kai, L.; Guo, K.; Zhao, W. Isolation and characterization of a new bacterium capable of biotransforming cis-epoxysuccinic acid to d(−) -tartaric acid. FEMS Microbiol. Lett. 2007, 267, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Xie, Z.; Bao, W.; Zhang, J. Isolation and identification of a novel cis-epoxysuccinate hydrolase producing Bordetella sp. BK-52 and optimization of enzyme production. Acta Microbiol. Sin. 2008, 48, 1075–1081. [Google Scholar]
- Wang, Z.; Wang, Y.; Su, Z. Purification and characterization of a cis-epoxysuccinic acid hydrolase from Nocardia tartaricans CAS-52, and expression in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, L.; Pan, H.; Bao, W.; Sun, W.; Xie, Z.; Zhang, J.; Zhao, Y. Purification and characterization of a novel cis-epoxysuccinate hydrolase from Klebsiella sp. that produces l(+)-tartaric acid. Biotechnol. Lett. 2014, 36, 2325–2330. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Pan, H.; Zhang, Z.; Cheng, Y.; Xie, Z.; Zhang, J. Isolation of the stable strain Labrys sp. BK-8 for l(+)-tartaric acid production. J. Biosci. Bioeng. 2015, 119, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Asai, Y.; Kobayashi, M.; Uchida, K.; Terasawa, M. DNA encoding cis-epoxysuccinic acid hydrolase and its utilization. Jp. Patent 2000-295992, 24 October 2000. CAN 133:307122. [Google Scholar]
- Cui, G.Z.; Wang, S.; Li, Y.; Tian, Y.J.; Feng, Y.; Cui, Q. High yield recombinant expression, characterization and homology modeling of two types of cis-epoxysuccinic acid hydrolases. Protein J. 2012, 31, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Vasu, V.; Kumaresan, J.; Babu, M.G.; Meenakshisundaram, S. Active site analysis of cis-epoxysuccinate hydrolase from Nocardia tartaricans using homology modeling and site-directed mutagenesis. Appl. Microbiol. Biotechnol. 2012, 93, 2377–2386. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Pan, H.; Bao, W.; Sun, W.; Xie, Z.; Zhang, J.; Zhao, Y. Cloning, homology modeling, and reaction mechanism analysis of a novel cis-epoxysuccinate hydrolase from Klebsiella sp. Biotechnol. Lett. 2014, 36, 2537–2544. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Xie, Z.; Bao, W.; Cheng, Y.; Zhang, J.; Li, Y. Site-directed mutagenesis of epoxide hydrolase to probe catalytic amino acid residues and reaction mechanism. FEBS Lett. 2011, 585, 2545–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, W.; Pan, H.; Zhang, Z.; Cheng, Y.; Xie, Z.; Zhang, J.; Li, Y. Analysis of essential amino acid residues for catalytic activity of cis-epoxysuccinate hydrolase from Bordetella sp. BK-52. Appl. Microbiol. Biotechnol. 2014, 98, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Liu, X.; Cui, G.Z.; Cui, Q.; Wang, X.; Feng, Y. Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(−)-tartaric acid. Chem. Commun. 2018, 54, 8482–8485. [Google Scholar] [CrossRef] [PubMed]
- Qiao, P.; Wu, M.; Zhu, L.; Zhang, Y.; Yang, L.; Fei, H.; Lin, J. Enhancing the thermal tolerance of a cis-epoxysuccinate hydrolase via combining directed evolution with various semi-rational redesign methods. J. Mol. Catal. B Enzym. 2015, 121, 96–103. [Google Scholar] [CrossRef]
- Zhang, C.; Pan, H.; Yao, L.; Bao, W.; Wang, J.; Xie, Z.; Zhang, J. Single point mutations enhance activity of cis-epoxysuccinate hydrolase. Biotechnol. Lett. 2016, 38, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Bučko, M.; Vikartovská, A.; Lacík, I.; Kolláriková, G.; Gemeiner, P.; Pätoprstý, V.; Brygin, M. Immobilization of a whole-cell epoxide-hydrolyzing biocatalyst in sodium alginate−cellulose sulfate−poly(methylene-co-guanidine) capsules using a controlled encapsulation process. Enzyme Microb. Technol. 2005, 36, 118–126. [Google Scholar] [CrossRef]
- Rosenberg, M.; Miková, H.; Krištofíková, L. Production of l-tartaric acid by immobilized bacterial cells Nocardia tartaricans. Biotechnol. Lett. 1999, 21, 491–495. [Google Scholar] [CrossRef]
- Kurillová, L.; Gemeiner, P.; Vikartovská, A.; Miková, H.; Rosenberg, M.; Ilavský, M. Calcium pectate gel beads for cell entrapment. 6. Morphology of stabilized and hardened calcium pectate gel beads with cells for immobilized biotechnology. J. Microencaps. 2000, 17, 279–296. [Google Scholar]
- Zhang, J.-G.; Qian, Y.-J. Production of l(+)-tartaric acid by immobilized Corynebacterium sp. JZ-1. Chin. J. Biotechnol./Shengwu Gongcheng Xuebao 2000, 16, 188–192. [Google Scholar]
- Vikartovská, A.; Bučko, M.; Gemeiner, P.; Nahálka, J.; Pätoprstý, V.; Hrabárová, E. Flow calorimetry—A useful tool for determination of immobilized cis-epoxysuccinate hydrolase activity from Nocardia tartaricans. Artif. Cells Blood Substit. Biotechnol. 2004, 32, 77–89. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Shi, H.; Su, Z. Improvement of the production efficiency of l-(+)-tartaric acid by heterogeneous whole-cell bioconversion. Appl. Biochem. Biotechnol. 2014, 172, 3989–4001. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cui, G.Z.; Song, X.F.; Feng, Y.; Cui, Q. Efficiency and stability enhancement of cis-epoxysuccinic acid hydrolase by fusion with a carbohydrate binding module and immobilization onto cellulose. Appl. Biochem. Biotechnol. 2012, 168, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Bao, W.; Pan, H.; Xie, Z.; Zhang, J. Production of l(+)-tartaric acid by immobilized Rhizobium strain BK-20. Chin. J. Biotechnol./Shengwu Gongcheng Xuebao 2014, 32, 315–319. [Google Scholar]
- Jin, T.-L.; Zhang, P.-D. Studies on increasing turnover rate of endoenzyme of immobilized cells by permealilizing and crosslinking method. Ind. Microbiol. 2003, 33, 14–19, 22. [Google Scholar]
- Dong, W.; Zhao, F.; Xin, F.; He, A.; Zhang, Y.; Wu, H.; Fang, Y.; Zhang, W.; Ma, J.; Jiang, M. Ultrasound-assisted D-tartaric acid whole-cell bioconversion by recombinant Escherichia coli. Ultrason. Sonochem. 2018, 42, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Su, M.; Li, Y.; Wang, Y.; Su, Z. Production of tartaric acid using immobilized recominant cis-epoxysuccinate hydrolase. Biotechnol. Lett. 2017, 39, 1859–1863. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Shi, H.; Su, Z. Expression and production of recombinant cis-epoxysuccinate hydrolase in Escherichia coli under the control of temperature-dependent promoter. J. Biotechnol. 2012, 162, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Widersten, M.; Gurell, A.; Lindberg, D. Structure-function relationships of epoxide hydrolases and their potential use in biocatalysis. Biochim. Biophys. Acta 2010, 1800, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Arand, M.; Hallberg, B.M.; Zou, J.; Bergfors, T.; Oesch, F.; van der Werf, M.J.; de Bont, J.A.M.; Jones, T.A.; Mowbray, S.L. Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site. EMBO J. 2003, 22, 2583–2592. [Google Scholar] [CrossRef] [PubMed]
- Fillgrove, K.L.; Pakhomova, S.; Schaab, M.R.; Newcomer, M.E.; Armstrong, R.N. Structure and mechanism of the genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes. Biochemistry 2007, 46, 8110–8120. [Google Scholar] [CrossRef] [PubMed]

| CESH | Genus | Species | Gene | Reference |
|---|---|---|---|---|
| CESH[L] | Acetobacter | Acetobacter curtus | [26] | |
| Acinetobacter | Acinetobacter tartarogenes | [23] | ||
| Agrobacterium | Agrobacterium aureum | [22] | ||
| Agrobacterium viscosum | [22] | |||
| Corynebacterium | Corynebacterium S-13 | [26] | ||
| Corynebacterium sp. JZ-1 | [28] | |||
| Klebsiella | Klebsiella sp. BK-58 | KF977193 | [38,43] | |
| Labrys | Labrys sp. BK-8 | [39] | ||
| Nocardia | Nocardia tartaricans | [24] | ||
| Nocardia tartaricans SW13-57 | [29] | |||
| Nocardia tartaricans CAS-52 | JQ267565 | [37] | ||
| Pseudomonas | Pseudomonas sp. KB-86 | [22] | ||
| Rhizobium | Rhizobium validum | [22,27] | ||
| Rhodococcus | Rhodococcus opacus | DQ471957 | [17] | |
| Rhodococcus ruber M1 | [34] | |||
| Rhodococcus rhodochrous | [3] | |||
| CESH[D] | Achromobacter | Achromobacter tartarogenes | [21] | |
| Achromobacter epoxylyticus | [21] | |||
| Achromobacter acinus | [21] | |||
| Achromobacter sericatus | [21] | |||
| Alcaligenes | Alcaligenes epoxylyticus | [21] | ||
| Alcaligenes margaritae | [21] | |||
| Alcaligenes sp. MCI3611 | 1 | [33,40] | ||
| Bordetella | Bordetella sp. strain 1–3 | [35] | ||
| Bordetella sp. BK-52 | EU053208 | [19,36] | ||
| Pseudomonas | Pseudomonas putida | [31] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xuan, J.; Feng, Y. Enantiomeric Tartaric Acid Production Using cis-Epoxysuccinate Hydrolase: History and Perspectives. Molecules 2019, 24, 903. https://doi.org/10.3390/molecules24050903
Xuan J, Feng Y. Enantiomeric Tartaric Acid Production Using cis-Epoxysuccinate Hydrolase: History and Perspectives. Molecules. 2019; 24(5):903. https://doi.org/10.3390/molecules24050903
Chicago/Turabian StyleXuan, Jinsong, and Yingang Feng. 2019. "Enantiomeric Tartaric Acid Production Using cis-Epoxysuccinate Hydrolase: History and Perspectives" Molecules 24, no. 5: 903. https://doi.org/10.3390/molecules24050903
APA StyleXuan, J., & Feng, Y. (2019). Enantiomeric Tartaric Acid Production Using cis-Epoxysuccinate Hydrolase: History and Perspectives. Molecules, 24(5), 903. https://doi.org/10.3390/molecules24050903