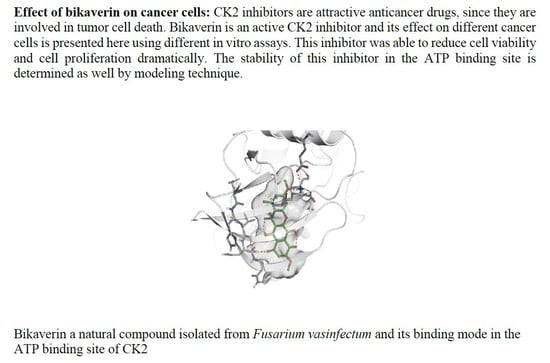
In Vitro and In Silico Evaluation of Bikaverin as a Potent Inhibitor of Human Protein Kinase CK2
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Compounds
3.2. Caco-2 Cell Permeability Assay
3.3. Cell Culture and Proliferation
3.4. Cell Viability Assay
3.5. IncuCyte® Cytotoxicity Assay
3.6. Molecular Dynamics
3.6.1. Computational Study
3.6.2. Protein Structure Preparation and Molecular Docking
3.6.3. Molecular Dynamics Simulation
3.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cozza, G.; Pinna, L.A. Casein kinases as potential therapeutic targets. Expert Opin. Ther. Targets 2016, 20, 319–340. [Google Scholar] [CrossRef]
- Ivanov, K.I.; Puustinen, P.; Gabrenaite, R.; Vihinen, H.; Ronnstrand, L.; Valmu, L.; Kalkkinen, N.; Makinen, K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell 2003, 15, 2124–2139. [Google Scholar] [CrossRef] [PubMed]
- Axtell, R.C.; Xu, L.; Barnum, S.R.; Raman, C. CD5-CK2 binding/activation-deficient mice are resistant to experimental autoimmune encephalomyelitis: Protection is associated with diminished populations of IL-17-expressing T cells in the central nervous system. J. Immunol. 2006, 177, 8542–8549. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, I.; Wang, H.X.; Feng, X.; Alenina, N.; Bader, M.; Prabhakar, B.S.; Li, P.F. Down-regulation of catalase and oxidative modification of protein kinase CK2 lead to the failure of apoptosis repressor with caspase recruitment domain to inhibit cardiomyocyte hypertrophy. J. Biol. Chem. 2008, 283, 5996–6004. [Google Scholar] [CrossRef] [PubMed]
- Jabor Gozzi, G.; Bouaziz, Z.; Winter, E.; Daflon-Yunes, N.; Aichele, D.; Nacereddine, A.; Marminon, C.; Valdameri, G.; Zeinyeh, W.; Bollacke, A.; et al. Converting potent indeno[1,2-b]indole inhibitors of protein kinase CK2 into selective inhibitors of the breast cancer resistance protein ABCG2. J. Med. Chem. 2015, 58, 265–277. [Google Scholar]
- Cozza, G. The Development of CK2 Inhibitors: From Traditional Pharmacology to in Silico Rational Drug Design. Pharmaceuticals 2017, 10, 26. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Drygin, D.; Streiner, N.; Chua, P.; Pierre, F.; O’Brien, S.E.; Bliesath, J.; Omori, M.; Huser, N.; Ho, C.; et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–11098. [Google Scholar]
- Baier, A.; Nazaruk, J.; Galicka, A.; Szyszka, R. Inhibitory influence of natural flavonoids on human protein kinase CK2 isoforms: Effect of the regulatory subunit. Mol. Cell Biochem. 2018, 444, 35–42. [Google Scholar] [CrossRef]
- Haidar, S.; Bouaziz, Z.; Marminon, C.; Laitinen, T.; Poso, A.; Le Borgne, M.; Jose, J. Development of Pharmacophore Model for Indeno[1,2-b]indoles as Human Protein Kinase CK2 Inhibitors and Database Mining. Pharmaceuticals 2017, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Li-Weber, M. Targeting apoptosis pathways in cancer by Chinese medicine. Cancer Lett. 2013, 332, 304–312. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE), 201308 version; C.C.G.I.: Montreal, QC, Canada, 2013.
- Kreitman, G.; Nord, F.F. Lycopersin, a pigment from Fusarium lycopersici. Arch. Biochem. 1949, 21, 457. [Google Scholar] [PubMed]
- Limon, M.C.; Rodriguez-Ortiz, R.; Avalos, J. Bikaverin production and applications. Appl. Microbiol. Biotechnol. 2010, 87, 21–29. [Google Scholar] [CrossRef]
- Kovac, L.; Bohmerova, E.; Fuska, J. Inhibition of mitochondrial functions by the antibiotics, bikaverin and duclauxine. J. Antibiot. 1978, 31, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Lale, G.J.; Gadre, R.V. Production of bikaverin by a Fusarium fujikuroi mutant in submerged cultures. AMB Express 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Arndt, B.; Studt, L.; Wiemann, P.; Osmanov, H.; Kleigrewe, K.; Kohler, J.; Krug, I.; Tudzynski, B.; Humpf, H.U. Genetic engineering, high resolution mass spectrometry and nuclear magnetic resonance spectroscopy elucidate the bikaverin biosynthetic pathway in Fusarium fujikuroi. Fungal Genet. Biol. 2015, 84, 26–36. [Google Scholar] [CrossRef]
- Deshmukh, R.; Purohit, H.J. siRNA mediated gene silencing in Fusarium sp. HKF15 for overproduction of bikaverin. Bioresour Technol. 2014, 157, 368–371. [Google Scholar] [CrossRef]
- Boyer, J.; Brown, D.; Liu, R.H. Uptake of quercetin and quercetin 3-glucoside from whole onion and apple peel extracts by Caco-2 cell monolayers. J. Agric. Food Chem. 2004, 52, 7172–7179. [Google Scholar] [CrossRef]
- Mao, X.; Wu, L.F.; Zhao, H.J.; Liang, W.Y.; Chen, W.J.; Han, S.X.; Qi, Q.; Cui, Y.P.; Li, S.; Yang, G.H.; et al. Transport of Corilagin, Gallic Acid, and Ellagic Acid from Fructus Phyllanthi Tannin Fraction in Caco-2 Cell Monolayers. Evid. Based Complement. Alternat Med. 2016, 9205379, 21. [Google Scholar] [CrossRef]
- Butkevich, A.N.; Mitronova, G.Y.; Sidenstein, S.C.; Klocke, J.L.; Kamin, D.; Meineke, D.N.H.; D’Este, E.; Kraemer, P.-T.; Danzl, J.G.; Belov, V.N.; et al. Fluorescent Rhodamines and Fluorogenic Carbopyronines for Super-Resolution STED Microscopy in Living Cells. Angew Chem. Int. Ed. Engl. 2016, 55, 3290–3294. [Google Scholar] [CrossRef]
- Artursson, P.; Palm, K.; Luthman, K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv. Drug Deliv. Rev. 2001, 46, 27–43. [Google Scholar] [CrossRef]
- Alchab, F.; Ettouati, L.; Bouaziz, Z.; Bollacke, A.; Delcros, J.G.; Gertzen, C.G.; Gohlke, H.; Pinaud, N.; Marchivie, M.; Guillon, J.; et al. Synthesis, Biological Evaluation and Molecular Modeling of Substituted Indeno[1,2-b]indoles as Inhibitors of Human Protein Kinase CK2. Pharmaceuticals 2015, 8, 279–302. [Google Scholar] [CrossRef]
- Nacereddine, A.; Bollacke, A.; Róka, E.; Marminon, C.; Bouaziz, Z.; Fenyvesi, F.; Bácskay, I.K.; Jose, J.; Perret, F.; Le Borgne, M. Self-Assembled Supramolecular Nanoparticles Improve the Cytotoxic Efficacy of CK2 Inhibitor THN7. Pharmaceuticals 2018, 11, 10. [Google Scholar] [CrossRef]
- Borowiecki, P.; Wawro, A.M.; Wińska, P.; Wielechowska, M.; Bretner, M. Synthesis of novel chiral TBBt derivatives with hydroxyl moiety. Studies on inhibition of human protein kinase CK2alpha and cytotoxicity properties. Eur. J. Med. Chem. 2014, 84, 364–374. [Google Scholar] [CrossRef]
- Li, D.; Chen, L.; Hu, Z.; Li, H.; Li, J.; wie, C.; Huang, Y.; Song, H.; Fang, L. Alterations of microRNAs are associated with impaired growth of MCF-7 breast cancer cells induced by inhibition of casein kinase 2. Int. J. Clin. Exp. Pathol. 2014, 7, 4008–4015. [Google Scholar]
- Salic, A.; Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef]
- Zhan, J.; Burns, A.M.; Liu, M.X.; Faeth, S.H.; Gunatilaka, A.L. Search for cell motility and angiogenesis inhibitors with potential anticancer activity: Beauvericin and other constituents of two endophytic strains of Fusarium oxysporum. J. Nat. Prod. 2007, 70, 227–232. [Google Scholar] [CrossRef]
- Fuska, J.; Proksa, B.; Fuskova, A. New potential cytotoxic and antitumor substances I. In vitro effect of bikaverin and its derivatives on cells of certain tumors. Neoplasma 1975, 22, 335–338. [Google Scholar]
- Kjaer, D.; Kjaer, A.; Pedersen, C.; Bu’lock, J.D.; Smith, J.R. Bikaverin and norbikaverin, benzoxanthentrione pigments of Gibberella fujikuroi. J. Chem. Soc. Perkin 1971, 16, 2792–2797. [Google Scholar] [CrossRef]
- Hubatsch, I.; Ragnarsson, E.G.; Artursson, P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nat. Protoc. 2007, 2, 2111–2119. [Google Scholar] [CrossRef]
- Artursson, P.; Karlsson, J. Correlation between oral drug absorption in humans and apparent drug permeability coefficients in human intestinal epithelial (Caco-2) cells. Biochem. Biophys. Res. Commun. 1991, 175, 880–885. [Google Scholar] [CrossRef]
- Tóth, Z.A.; Raatikainen, O.; Naaranlahti, T.; Auriola, S. Isolation and determination of alizarin in cell cultures of Rubia tinctorum and emodin in Dermocybe sanguinea using solid-phase extraction and high-performance liquid chromatography. J. Chromatogr. A 1993, 630, 423–428. [Google Scholar]
- Bischoff, N.; Raaf, J.; Olsen, B.; Bretner, M.; Issinger, O.G.; Niefind, K. Enzymatic activity with an incomplete catalytic spine: Insights from a comparative structural analysis of human CK2alpha and its paralogous isoform CK2alpha’. Mol. Cell Biochem. 2011, 356, 57–65. [Google Scholar] [CrossRef]
- Xue, L.Y.; Chiu, S.M.; Oleinick, N.L. Staurosporine-induced death of MCF-7 human breast cancer cells: A distinction between caspase-3-dependent steps of apoptosis and the critical lethal lesions. Exp. Cell. Res. 2003, 283, 135–145. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Small-Molecule Drug Discovery Suite 2018-1; Schrödinger, LLC: New York, NY, USA, 2018.
- Epik, S. LLC, New York, NY, USA, 2016; Impact, Schrödinger, LLC, New York, NY, USA, 2016; Prime, Schrödinger, LLC, New York, NY, USA, 2018.
- Glide, S. LLC, New York, NY, USA, 2016; Prime, Schrödinger, LLC, New York, NY, USA, 2018.
- Desmond Molecular Dynamics System, D.E.S.R.: New York, NY, USA, 2018. Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2018.
Sample Availability: Samples of the compounds are not available from the authors. |











© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haidar, S.; Aichele, D.; Birus, R.; Hielscher, J.; Laitinen, T.; Poso, A.; Jose, J. In Vitro and In Silico Evaluation of Bikaverin as a Potent Inhibitor of Human Protein Kinase CK2. Molecules 2019, 24, 1380. https://doi.org/10.3390/molecules24071380
Haidar S, Aichele D, Birus R, Hielscher J, Laitinen T, Poso A, Jose J. In Vitro and In Silico Evaluation of Bikaverin as a Potent Inhibitor of Human Protein Kinase CK2. Molecules. 2019; 24(7):1380. https://doi.org/10.3390/molecules24071380
Chicago/Turabian StyleHaidar, Samer, Dagmar Aichele, Robin Birus, Janine Hielscher, Tuomo Laitinen, Antti Poso, and Joachim Jose. 2019. "In Vitro and In Silico Evaluation of Bikaverin as a Potent Inhibitor of Human Protein Kinase CK2" Molecules 24, no. 7: 1380. https://doi.org/10.3390/molecules24071380
APA StyleHaidar, S., Aichele, D., Birus, R., Hielscher, J., Laitinen, T., Poso, A., & Jose, J. (2019). In Vitro and In Silico Evaluation of Bikaverin as a Potent Inhibitor of Human Protein Kinase CK2. Molecules, 24(7), 1380. https://doi.org/10.3390/molecules24071380