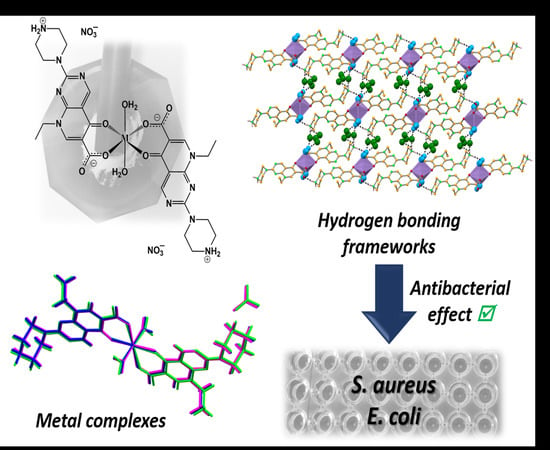
Bioactivity of Isostructural Hydrogen Bonding Frameworks Built from Pipemidic Acid Metal Complexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural Characterisation
Shelf and Thermal Stability of the Compounds
2.2. Antibacterial Activity Assays
2.3. General Toxicity Assay
3. Experimental Section
3.1. Reagents
3.2. Metal Complexes Synthesis
3.3. Single Crystal X-ray Diffraction (SCXRD)
3.4. Powder X-ray Diffraction (PXRD)
3.5. Variable Temperature Powder X-ray Diffraction (VT-PXRD)
3.6. Hot-Stage Microscopy (HSM)
3.7. Differential Scanning Calorimetry (DSC) and Thermogravimetric Analysis (TGA)
3.8. Infrared Spectroscopy (IR)
3.9. Nuclear Magnetic Resonance Spectroscopy (NMR)
3.10. Antibacterial Activity Assays
3.11. General Toxicity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Andersson, M.I.; MacGowan, A.P. Development of the quinolones. J. Antimicrob. Chemother. 2003, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Uivarosi, V. Metal Complexes of Quinolone Antibiotics and Their Applications: An Update. Molecules 2013, 18, 11153–11197. [Google Scholar] [CrossRef] [PubMed]
- Daneshtalab, M.; Ahmed, A. Nonclassical biological activities of quinolone derivatives. J. Pharm. Pharm. Sci. 2012, 15, 52–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sissi, C.; Palumbo, M. The quinolone family: From antibacterial to anticancer agents. Curr. Med. Chem. Agents 2003, 3, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA Topoisomerases and Their Poisoning by Anticancer and Antibacterial Drugs. Chem. Boil. 2010, 17, 421–433. [Google Scholar] [CrossRef] [Green Version]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef] [Green Version]
- Ball, P. Quinolone generations: Natural history or natural selection? J. Antimicrob. Chemother. 2000, 46, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of Quinolone Action and Resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Noble, C.G.; Barnard, F.M.; Maxwell, A. Quinolone-DNA Interaction: Sequence-Dependent Binding to Single-Stranded DNA Reflects the Interaction within the Gyrase-DNA Complex. Antimicrob. Agents Chemother. 2003, 47, 854–862. [Google Scholar] [CrossRef] [Green Version]
- Barrett, J.; Bernstein, J.; Krause, H.; Hilliard, J.; Ohemeng, K. Testing Potential Gyrase Inhibitors of Bacterial DNA Gyrase: A Comparison of the Supercoiling Inhibition Assay and “Cleavable Complex” Assay. Anal. Biochem. 1993, 214, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, J.; Minami, S. Pyrido [2,3-d]pyrimidine antibacterial agents. 3. 8-Alkyl- and 8-vinyl-5,8-dihydro-5-oxo-2-(1-piperazinyl)pyrido[2,3-d]pyrimidine-6-carboxylic acids and their derivatives. J. Med. Chem. 1975, 18, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, I.; Martínez-Carrera, S.; Garciá-Blanco, S. Strucure of pipemidic acid. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1986, 42, 1618–1621. [Google Scholar] [CrossRef]
- Appelbaum, P.C.; Hunter, P. The fluoroquinolone antibacterials: Past, present and future perspectives. Int. J. Antimicrob. Agents 2000, 16, 5–15. [Google Scholar] [CrossRef]
- Van Oosterom, R.A.A.; Hartman, E.G. Pipemidic acid, a new treatment for recurrent urinary tract infection in small animals. Veter Q. 1986, 8, 2–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirai, K.; Ito, A.; Abe, Y.; Suzue, S.; Irikura, T.; Inoue, M.; Mitsuhashi, S. Comparative Activities of AM-715 and Pipemidic and Nalidixic Acids Against Experimentally Induced Systemic and Urinary Tract Infections. Antimicrob. Agents Chemother. 1981, 19, 188–189. [Google Scholar] [CrossRef] [Green Version]
- Lavorgna, M.; Iacovino, R.; Russo, C.; Di Donato, C.; Piscitelli, C.; Isidori, M. A New Approach for Improving the Antibacterial and Tumor Cytotoxic Activities of Pipemidic Acid by Including It in Trimethyl-β-cyclodextrin. Int. J. Mol. Sci. 2019, 20, 416. [Google Scholar] [CrossRef] [Green Version]
- Turel, I. The interactions of metal ions with quinolone antibacterial agents. Coord. Chem. Rev. 2002, 232, 27–47. [Google Scholar] [CrossRef]
- Singh, R.; Debnath, A.; Masram, D.T.; Rathore, D. ChemInform Abstract: Synthesis and Biological Activities of Selected Quinolone-Metal Complexes. Res. J. Chem. Sci. 2015, 46, 83–94. [Google Scholar] [CrossRef]
- Andre, V.; Galego, F.; Martins, M. Mechanochemical Assembly of Nalidixic Acid Bioinspired Metal–Organic Compounds and Complexes toward Improved Solubility. Cryst. Growth Des. 2018, 18, 2067–2081. [Google Scholar] [CrossRef]
- Andre, V.; Da Silva, A.R.F.; Fernandes, A.; Frade, R.; Garcia, C.; Rijo, P.; Antunes, A.M.M.; Rocha, J.; Duarte, M.T. Mg- and Mn-MOFs Boost the Antibiotic Activity of Nalidixic Acid. ACS Appl. Bio Mater. 2019, 2, 2347–2354. [Google Scholar] [CrossRef]
- Efthimiadou, E.K.; Sanakis, Y.; Katsaros, N.; Karaliota, A.; Psomas, G. Transition metal complexes with the quinolone antibacterial agent pipemidic acid: Synthesis, characterization and biological activity. Polyhedron 2007, 26, 1148–1158. [Google Scholar] [CrossRef]
- Yang, L.; Tao, D.; Yang, X.; Li, Y.; Guo, Y. Synthesis, characterization, and antibacterial activities of some rare Earth metal complexes of pipemidic acid. Chem. Pharm. Bull. 2003, 51, 494–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groom, C.; Allen, F.H. The Cambridge Structural Database in Retrospect and Prospect. Angew. Chem. Int. Ed. 2014, 53, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Xing, G.; Zhang, Y. Tuning structural dimensionalities of two new luminescent Cd(II) compounds: Different dicarboxylate coligands. J. Mol. Struct. 2016, 1123, 133–137. [Google Scholar] [CrossRef]
- Sha, J.-Q.; Li, X.; Sha, J.-Q.; Zhang, Y.-H.; Yan, H. Nickel complexes of the different quinolone antibacterial drugs: Synthesis, structure and interaction with DNA. Inorg. Chim. Acta 2012, 383, 178–184. [Google Scholar] [CrossRef]
- Xue, C.-M.; Li, S.-X.; Zhang, L.; Sha, J.-Q.; Zheng, T.-Y.; Zhang, Q.-N.; Li, L. Hydrothermal Synthesis, Characterization and Electrocatalytic/Photocatalytic Activities of New Polyoxometalate Based Hybrid Compound. J. Inorg. Organomet. Polym. Mater. 2013, 23, 1468–1476. [Google Scholar] [CrossRef]
- Li, M.-T.; Sun, J.-W.; Sha, J.-Q.; Wu, H.-B.; Zhang, E.-L.; Zheng, T.-Y. An unprecedented Ag–pipemidic acid complex with helical structure: Synthesis, structure and interaction with CT-DNA. J. Mol. Struct. 2013, 1045, 29–34. [Google Scholar] [CrossRef]
- Duan, L.-N.; Dang, Q.-Q.; Han, C.-Y.; Zhang, X.-M. An interpenetrated bioactive nonlinear optical MOF containing a coordinated quinolone-like drug and Zn(ii) for pH-responsive release. Dalton Trans. 2015, 44, 1800–1804. [Google Scholar] [CrossRef]
- He, J.-H.; Xiao, D.-R.; Yan, S.-W.; Sun, D.-Z.; Chen, H.; Wang, X.; Yang, J.; Ye, Z.-L.; Yuan, R.; Wang, E. A series of novel 1D coordination polymers constructed from metal–quinolone complex fragments linked by aromatic dicarboxylate ligands. Solid State Sci. 2012, 14, 1203–1210. [Google Scholar] [CrossRef]
- Ye, Z.-L.; Xin, G.-H.; Zhang, F.-T.; Xiao, D.-R. Diaqua(5-carboxybenzene-1,3-dicarboxylato-kO1) [8-ethyl-5-oxo-2-(piperazin-4-ium-1-yl)-5,8-dihydro-pyrido[2–d]pyrimidine-6-carboxylato-k2O5,O6] zinc monohydrate. Acta Crystallogr. Sect. E Struct. Rep. Online 2013, 69, m127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Zhu, D.-S.; Song, X.-D.; An, Z. Poly [[bis-[μ2-8-ethyl-5-oxo-2-(piperazin-1-yl)-5,8-dihydro-pyrido[2,3-d]pyrimidine-6-carboxylato]zinc(II)] dihydrate]. Acta Crystallogr. Sect. E Struct. Rep. Online 2009, 65, m1223. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hu, W.-P.; An, Z. Poly[[bis-[μ2-8-ethyl-5-oxo-2-(piperazin-1-yl)-5,8-dihydro-pyrido[2,3-d]pyrimidine-6-carboxylato]manganese(II)] dihydrate]. Acta Crystallogr. Sect. E Struct. Rep. Online 2008, 64, m547. [Google Scholar] [CrossRef]
- Zhu, L.; Zhou, T. Crystal structure of bis(8-ethyl-5-oxo-2-(piperazin-4-ium-1-yl)-5,8-dihydro-pyrido[2,3-d]pyrimidine-6-carboxylato-κ2O,O′) copper(II) benzene-1,4-dicarboxylate dihydrate, C36H42CuN10O12. Z. Für Krist. New Cryst. Struct. 2016, 231, 447. [Google Scholar] [CrossRef] [Green Version]
- He, J.-H.; Sun, D.-Z.; Xiao, D.-R.; Yan, S.-W.; Chen, H.; Wang, X.; Yang, J.; Wang, E. Syntheses and structures of five 1D coordination polymers based on quinolone antibacterial agents and aromatic polycarboxylate ligands. Polyhedron 2012, 42, 24–29. [Google Scholar] [CrossRef]
- Xiao, D.-R.; He, J.-H.; Sun, D.-Z.; Chen, H.; Yan, S.-W.; Wang, X.; Yang, J.; Yuan, R.; Wang, E. Three 3D Metal-Quinolone Complexes Based on Trimetallic or Rod-Shaped Secondary Building Units. Eur. J. Inorg. Chem. 2012, 2012, 1783–1789. [Google Scholar] [CrossRef]
- Sun, D.-Z.; Zhang, G.-J.; Chen, H.-Y.; He, J.-H.; Yan, S.-W. Diaquabis[8-ethyl-5-oxo-2-(piperazin-4-ium-1-yl)-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxylato] copper(II) bis[4-(4-carboxyphenoxy)benzoate]. Acta Crystallogr. Sec. E Struct. Rep. Online 2011, 67, m388. [Google Scholar] [CrossRef]
- Capiod, T.; Shuba, Y.; Skryma, R.; Prevarskaya, N. Calcium signalling and cancer cell growth. Membr. Biog. 2007, 45, 405–427. [Google Scholar] [CrossRef]
- Deshpande, C.N.; Ruwe, T.; Shawki, A.; Xin, V.; Vieth, K.R.; Valore, E.V.; Qiao, B.; Ganz, T.; Nemeth, E.; MacKenzie, B.; et al. Calcium is an essential cofactor for metal efflux by the ferroportin transporter family. Nat. Commun. 2018, 9, 3075. [Google Scholar] [CrossRef]
- Lacaz-Vieira, F. Calcium Site Specificity. J. Gen. Physiol. 1997, 110, 727–740. [Google Scholar] [CrossRef] [Green Version]
- Roth, J.; Ponzoni, S.; Aschner, M. Manganese Homeostasis and Transport. In Metallomics and the Cell; Banci, L., Ed.; Springer Netherlands: Dordrecht, The Netherland, 2013; pp. 169–201. [Google Scholar] [CrossRef]
- Frassinetti, S.; Bronzetti, G.L.; Caltavuturo, L.; Cini, M.; Della Croce, C. The role of zinc in life: A review. J. Environ. Pathol. Toxicol. Oncol. 2006, 25, 597–610. [Google Scholar] [CrossRef]
- Kaur, K.; Gupta, R.; Saraf, S.A.; Saraf, S.K. Zinc: The Metal of Life. Compr. Rev. Food Sci. Food Saf. 2014, 13, 358–376. [Google Scholar] [CrossRef]
- Andre, V.; Hardeman, A.; Halasz, I.; Stein, R.S.; Jackson, G.J.; Reid, D.G.; Duer, M.J.; Curfs, C.; Duarte, M.T.; Friščić, T. Mechanosynthesis of the Metallodrug Bismuth Subsalicylate from Bi2O3 and Structure of Bismuth Salicylate without Auxiliary Organic Ligands. Angew. Chem. Int. Ed. 2011, 50, 7858–7861. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.L.; Cao, Q.; Browne, D.L. Mechanochemistry as an emerging tool for molecular synthesis: What can it offer? Chem. Sci. 2018, 9, 3080–3094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quaresma, S.; Andre, V.; Fernandes, A.; Duarte, M.T. Mechanochemistry—A green synthetic methodology leading to metallodrugs, metallopharmaceuticals and bio-inspired metal-organic frameworks. Inorg. Chim. Acta 2017, 455, 309–318. [Google Scholar] [CrossRef]
- James, S.L.; Adams, C.J.; Bolm, C.; Braga, D.; Collier, P.; Friščić, T.; Grepioni, F.; Harris, K.D.M.; Hyett, G.; Jones, W.; et al. Mechanochemistry: Opportunities for new and cleaner synthesis. Chem. Soc. Rev. 2012, 41, 413–447. [Google Scholar] [CrossRef] [Green Version]
- Do, J.-L.; Friščić, T. Mechanochemistry: A Force of Synthesis. ACS Cent. Sci. 2016, 3, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Hasa, D.; Rauber, G.S.; Voinovich, D.; Jones, W. Cocrystal Formation through Mechanochemistry: From Neat and Liquid-Assisted Grinding to Polymer-Assisted Grinding. Angew. Chem. Int. Ed. 2015, 54, 7371–7375. [Google Scholar] [CrossRef]
- Marçalo, J.; Garcia, C.; Reis, C.P.; Teodósio, C.; Oliveira, C.; Oliveira, C.; Roberto, A. Biological activity screening of seven Plectranthus species. J. Biomed. Biopharm. Res. 2017, 14, 95–108. [Google Scholar] [CrossRef]
- Alanís-Garza, B.A.; González-González, G.; Salazar-Aranda, R.; De Torres, N.W.; Rivas-Galindo, V. Screening of antifungal activity of plants from the northeast of Mexico. J. Ethnopharmacol. 2007, 114, 468–471. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer17; University of Western Australia: Perth, Australia, 2017. [Google Scholar]
- Zhang, Y.; Mu, J.; Han, J.; Gu, X.-J. An improved brine shrimp larvae lethality microwell test method. Toxicol. Mech. Methods 2011, 22, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Rijo, P.; Molpeceres, J.; Figueiredo, I.V.; Ascensão, L.; Fernandes, A.S.; Roberto, A.; Reis, C.P. Polymeric nanoparticles modified with fatty acids encapsulating betamethasone for anti-inflammatory treatment. Int. J. Pharm. 2015, 493, 271–284. [Google Scholar] [CrossRef]
- Bruker. Bruker AXS: APEX3; Bruker Analytical Systems: Madison, WI, USA, 2016. [Google Scholar]
- Bruker. Bruker AXS: SAINT+ (6.22); Bruker Analytical Systems: Madison, WI, USA, 2014. [Google Scholar]
- Bruker. Bruker AXS: SADABS; Bruker Analytical Systems: Madison, WI, USA, 2014. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Macrae, C.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.E.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; Van De Streek, J.; Wood, P.A. Mercury CSD 2.0–new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Bruker. Bruker AXS: DIFFRAC.EVA V4.1; Bruker AXS GmbH: Karlsruhe, Germany, 2015. [Google Scholar]
- Siopa, F.; Figueiredo, T.; Frade, R.; Neto, I.; Meirinhos, A.; Reis, C.P.; Sobral, R.G.; Afonso, C.A.M.; Rijo, P. Choline-Based Ionic Liquids: Improvement of Antimicrobial Activity. Chemistryselect 2016, 1, 5909–5916. [Google Scholar] [CrossRef]
Sample Availability: Samples of the synthesised compounds are available from the authors. |









| Sym. Op. | D–H⋯A | d (D–H) (Å) | d (H⋯A) (Å) | d (D⋯A) (Å) | DĤA (°) | |
|---|---|---|---|---|---|---|
| I | x, y, z | N5–H1N⋯O4 | 0.91(4) | 2.43(5) | 3.065(5) | 126(4) |
| x, y, z | N5–H1N⋯O6 | 0.91(4) | 2.02(4) | 2.936(6) | 177(5) | |
| 1 − x, 2 − y, 1− z | O1w–H1w⋯O2 | 0.90(3) | 1.86(3) | 2.742(4) | 169(6) | |
| -½ + x, −½ + y, z | N5–H2N⋯O2 | 0.93(4) | 1.82(4) | 2.741(4) | 170(4) | |
| ½ + x, ½ − y, ½ + z | O1w–H2w⋯O5 | 0.89(6) | 2.17(5) | 2.990(6) | 153(6) | |
| ½ + x, ½ − y, ½ + z | O1w–H2w⋯O6 | 0.89(6) | 2.22(6) | 2.998(5) | 146(5) | |
| II | x, y, z | N5–H1N⋯O4 | 0.89(3) | 2.58(3) | 3.069(5) | 116(3) |
| x, y, z | N5–H1N⋯O6 | 0.89(3) | 2.06(3) | 2.933(5) | 168(3) | |
| x, −1 + y, z | O1w–H1w⋯O2 | 0.88(2) | 1.89(2) | 2.753(3) | 167(4) | |
| −½ + x, −½ + y, z | N5–H2N⋯O2 | 0.87(3) | 1.86(3) | 2.733(4) | 174(4) | |
| ½ − x, ½ + y, z | O1w–H2w⋯O5 | 0.87(3) | 2.17(3) | 2.974(5) | 154(3) | |
| ½ − x, ½ + y, z | O1w–H2w⋯O6 | 0.87(3) | 2.27(3) | 3.039(5) | 149(3) | |
| III | x, y, z | N5–H1N⋯O4 | 0.93(6) | 2.55(7) | 3.096(6) | 118(5) |
| x, y, z | N5–H1N⋯O6 | 0.93(6) | 2.03(6) | 2.952(6) | 172(7) | |
| 1 − x, 2 − y, 1 − z | O1w–H1w⋯O2 | 0.89(3) | 1.85(4) | 2.740(6) | 172(7) | |
| ½ + x, −½ + y, z | N5–H2N⋯O2 | 0.93(3) | 1.86(4) | 2.769(5) | 168(5) | |
| −½ + x, ½ − y, −½ + z | O1w–H2w⋯O5 | 0.89(6) | 2.43(7) | 3.031(7) | 125(6) | |
| −½ + x, ½ − y, −½ + z | O1w–H2w⋯O6 | 0.89(6) | 2.09(6) | 2.978(6) | 171(5) |
| Compounds | S. aureus | E. coli |
|---|---|---|
| MIC (µg/mL) | MIC (µg/mL) | |
| Pipemidic acid | 7.81 | 15.62 |
| Mn(NO3)2·4H2O | 3.90 | 62.50 |
| Complex I | 3.90 | 7.81 |
| Zn(NO3)2·6H2O | 31.25 | 31.25 |
| Complex II | 7.81 | 7.81 |
| Ca(NO3)2·4H2O | 7.81 | 62.50 |
| Complex III | 15.62 | 7.81 |
| Negative control (DMSO) | 62.50 | 62.50 |
| Positive control | 0.488 (VAN) | 0.488 (NOR) |
| Sample | % Viability | % Viability Range |
|---|---|---|
| Salt | 87.43 ± 3.77 | 83.66–91.20 |
| Sample solvent | 84.87 ± 5.70 | 79.17–90.57 |
| PA | 77.55 ± 2.00 | 75.55–79.55 |
| Complex I | 77.80 ± 2.09 | 75.71–79.89 |
| Complex II | 72.19 ± 5.93 | 66.26–78.12 |
| Complex III | 79.67 ± 4.07 | 75.60–83.74 |
| Negative control | 0 ± 0 | 0 |
| Compound | Complex I | Complex II | Complex III |
|---|---|---|---|
| Pipemidic acid | 0.1508 g | 0.1519 g | 0.1518 g |
| (0.50 mmol) | (0.50 mmol) | (0.50 mmol) | |
| M | 0.0646 g | 0.0778 g | 0.0687 g |
| (0.25 mmol) | (0.25 mmol) | (0.25 mmol) |
| I | II | III | |
|---|---|---|---|
| Formula | (C28H34N10O6Mn·2(H2O))·2(NO3) | (C28H34N10O6Zn·2(H2O))·2(NO3) | (C28H34N10O6Ca·2(H2O))·2(NO3) |
| Fw | 821.64 | 832.08 | 806.78 |
| Crystal form, colour | Plate, colourless | Block, colourless | Needle, colourless |
| Crystal size (mm) | 0.05 × 0.05 × 0.12 | 0.02 × 0.12 × 0.22 | 0.04 × 0.06 × 0.25 |
| Crystal system | Monoclinic | Monoclinic | Monoclinic |
| Space group | C2/c | C2/c | C2/c |
| a, Å | 24.347(5) | 24.282(3) | 24.533(7) |
| b, Å | 6.9370(14) | 6.8977(10) | 7.0158(18) |
| c, Å | 20.234(4) | 20.316(3) | 20.251(6) |
| β ° | 99.740(7) | 100.656(5) | 98.103(15) |
| Z | 4 | 4 | 4 |
| V, Å3 | 3368.1(12) | 3344.1(8) | 3450.8(16) |
| Dc, g cm−3 | 1.620 | 1.653 | 1.553 |
| μ(Mo Kα), mm−1 | 0.482 | 0.825 | 0.270 |
| θ range (°) | 2.869–26.576 | 2.406–26.578 | 2.445–26.556 |
| Refl. Collected/ | |||
| Independent refl. | 25703/3513 | 16919/3410 | 57149/3561 |
| Rint | 0.1696 | 0.1100 | 0.2113 |
| R1a, wR2b [I ≥ 2σ(I)] | 0.0574, 0.1268 | 0.0628, 0.1605 | 0.0733, 0.1801 |
| GOF on F2 | 1.039 | 1.017 | 1.103 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
C. Alves, P.; Rijo, P.; Bravo, C.; M. M. Antunes, A.; André, V. Bioactivity of Isostructural Hydrogen Bonding Frameworks Built from Pipemidic Acid Metal Complexes. Molecules 2020, 25, 2374. https://doi.org/10.3390/molecules25102374
C. Alves P, Rijo P, Bravo C, M. M. Antunes A, André V. Bioactivity of Isostructural Hydrogen Bonding Frameworks Built from Pipemidic Acid Metal Complexes. Molecules. 2020; 25(10):2374. https://doi.org/10.3390/molecules25102374
Chicago/Turabian StyleC. Alves, Paula, Patrícia Rijo, Catarina Bravo, Alexandra M. M. Antunes, and Vânia André. 2020. "Bioactivity of Isostructural Hydrogen Bonding Frameworks Built from Pipemidic Acid Metal Complexes" Molecules 25, no. 10: 2374. https://doi.org/10.3390/molecules25102374
APA StyleC. Alves, P., Rijo, P., Bravo, C., M. M. Antunes, A., & André, V. (2020). Bioactivity of Isostructural Hydrogen Bonding Frameworks Built from Pipemidic Acid Metal Complexes. Molecules, 25(10), 2374. https://doi.org/10.3390/molecules25102374