Anticancer Activity of Pyrimethamine via Ubiquitin Mediated Degradation of AIMP2-DX2
Abstract
:1. Introduction
2. Results
2.1. Identification of Pyrimethamine as a Suppressor of DX2 Level
2.2. Antiproliferative Effect of Pyrimethamine on Cancer Cells but not Normal Cells
2.3. DX2 Level-Dependent Antiproliferative Effect of Pyrimethamine
2.4. Specific Suppression of DX2 Level by Pyrimethamine
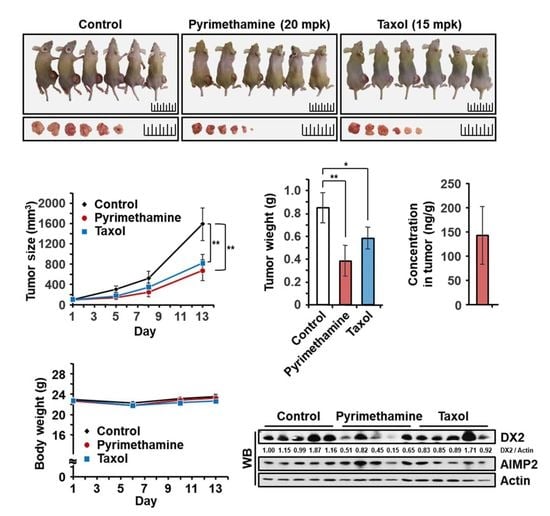
2.5. Inhibitory Effect of Pyrimethamine on Tumorigenesis In Vivo
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Materials
4.2. Library Screening Based on a Nanoluciferase Assay
4.3. Cell Viability Assay
4.4. Assessment of Cell Viability Using DX2- and EV (Empty Vector)-Inducible A549
4.5. Immunoblotting and RT-PC
4.6. Ubiquitination Assay
4.7. Animals
4.8. In Vivo Mouse Xenograft
4.9. Statistics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rajendran, V.; Kalita, P.; Shukla, H.; Kumar, A.; Tripathi, T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 2018, 111, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Park, S.G.; Ewalt, K.L.; Kim, S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: New perspectives on housekeepers. Trends Biochem. Sci. 2005, 30, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; You, S.; Hwang, D. Aminoacyl-tRNA synthetases and tumorigenesis: More than housekeeping. Nat. Rev. Cancer 2011, 11, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Maeng, S.J.; Cho, H.J.; Choi, Y.S.; Chung, J.M.; Lee, S.; Kim, H.K.; Kim, J.H.; Eom, C.Y.; Kim, Y.G.; et al. Assembly of multi-tRNA synthetase complex via heterotetrameric glutathione tranferase-homology domains. J. Biol. Chem. 2015, 290, 29313–29328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.J.; Park, B.-J.; Kang, Y.-S.; Kim, H.J.; Park, J.-H.; Kang, J.W.; Lee, S.W.; Han, J.M.; Lee, H.-W.; Kim, S. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat. Genet. 2003, 34, 330–336. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, D.G.; Park, M.C.; Um, J.Y.; Han, J.M.; Park, S.G.; Choi, E.C.; Kim, S. AIMP2 promotes TNF-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J. Cell Sci. 2009, 122, 2710–2715. [Google Scholar] [CrossRef] [Green Version]
- Han, J.M.; Park, B.-J.; Park, S.G.; Oh, Y.S.; Choi, S.J.; Lee, S.W.; Hwang, S.-K.; Chang, S.-H.; Cho, M.-H.; Kim, S. AMIP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genetoxic stresses via p53. Proc. Natl. Acad. Sci. USA 2008, 105, 11206–11211. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.W.; Um, J.Y.; Kundu, J.K.; Surh, Y.J.; Kim, S.K. Multidirectional tumor-suppressive activity of AIMP2/p38 and the enhanced susceptibility of AIMP2 heterozygous mice to carcinogenesis. Carcinogenesis 2009, 30, 1638–1644. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.W.; Kim, D.G.; Lee, A.E.; Kim, H.R.; Lee, J.Y.; Kwon, N.H.; Shin, Y.K.; Hwang, S.K.; Chang, S.H.; Cho, M.H.; et al. Cancer-associated splicing variant of tumor suppressor AIMP2/p38: Pathological implication in tumorigenesis. PLoS Genet. 2011, 7, e1001351. [Google Scholar] [CrossRef]
- Choi, J.W.; Lee, J.W.; Kim, J.K.; Jeon, H.K.; Choi, J.J.; Kim, D.G.; Kim, B.G.; Nam, D.H.; Kim, H.J.; Yun, S.H.; et al. Splicing variant of AIMP2 as an effective target against chemoresistant ovarian cancer. J. Mol. Cell. Biol. 2012, 4, 164–173. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.S.; Kim, D.G.; Oh, Y.S.; Kwon, N.H.; Lee, J.Y.; Kim, D.; Park, S.-H.; Song, J.-H.; Lee, S.; Han, J.M.; et al. Chemical suppression of an oncogenic splicing variant of AIMP2 induces tumor regression. Biochem. J. 2013, 454, 411–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.chembank.org (accessed on 15 May 2020).
- Montoya, J.G.; Lisenfeld, O. Tpxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef]
- Hopper, A.T.; Brockman, A.; Wise, A.; Gould, J.; Barks, J.; Radke, J.B.; Sibley, L.D.; Zou, Y.; Thomas, S. Discovery of selective Toxoplasma gondii diyydrofolate reductase inhibitors for treatment of toxoplasmosis. J. Med. Chem. 2019, 62, 1562–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hujisduijnen, R.H.; Guy, R.K.; Chibale, K.; Haynes, R.K.; Peitz, I.; Kelter, G.; Phillips, M.A.; Vennerstorm, J.L.; Yuthavong, Y. Anticancer properties of distinct antimalarial drug classes. PLoS ONE 2013, 8, e82962. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Osman, I.; Orlow, S.J. Amtifolate activity of pyrimethamine enhances temozomide-induced cytotoxicity in melanoma cells. Mol. Cancer Res. 2009, 7, 703–712. [Google Scholar] [CrossRef] [Green Version]
- Giammarioli, A.M.; Maselli, A.; Casagrande, A.; Gambardella, L.; Gallina, A.; Spada, M.; Giovannetti, A.; Proietti, E.; Walter, M. Pyrimethamine induces apoptosis of melanoma cells via a caspase and cathepsin double edged mechanism. Cancer Res. 2008, 68, 5291–5300. [Google Scholar] [CrossRef] [Green Version]
- Dai, C.; Zhang, B.; Liu, X.; Guo, K.; Ma, S.; Cai, F.; Yang, Y.; Yao, Y.; Feng, M.; Bao, X.; et al. Pyrimethamine sensitizes pituitary adenomas cells to temozolamide through cathepsn B-dependent and caspase dependent pathways. Int. J. Cancer 2013, 133, 1982–1993. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.-X.; Lin, S.-H.; Lin, C.-C.; Yang, C.-C.; Yuan, S.-Y. In vitro and in vivo antitumor effects of pyrimethamine on non-small cell lung cancers. Anticancer Res. 2018, 38, 3435–3445. [Google Scholar] [CrossRef]
- Liu, H.; Qin, Y.; Zhai, D.; Zhang, Q.; Gu, J.; Tang, Y.; Yang, J.; Li, K.; Yang, L.; Chen, S.; et al. Antimalarial drug pyrimethamine plays a dual role in antitumor proliferation and metastasis through targeting DHFR and TP. Mol. Cancer Res. 2019, 18, 541–555. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.W.; Saadalla, A.; Ewida, A.H.; Al-Katranji, K.; Al-Saoudi, G.; Giaccone, Z.T.; Gounari, F.; Zhang, M.; Frank, D.A.; Khazai, K. The STAT3 inhibitor pyrimethamine displays anti-cancer and immune stimulatory effects in murine models of breast cancer. Cancer Immunol. Immunother. 2018, 67, 13–23. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Wang, H. Pyrimethamine Exerts significant antitumor effects on human ovarian cancer cells both in vitro and in vivo. Anticancer Drugs 2019, 30, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.-J.; Yan, L.; Zhang, J.; Zhang, W.-D. STAT3 as a potential therapeutic target in triple negative breast cancer: A systematic review. J. Exp. Clin. Res. 2019, 38, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Cho, H.Y.; Kim, D.G.; Roh, Y.; Son, S.; Mushtaq, A.U.; Kim, M.; Bhattarai, D.; Sivaraman, A.; Lee, Y.; et al. Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development. Nat. Chem. Biol. 2020, 16, 31–41. [Google Scholar] [CrossRef]
- Huang, S.-T.; Wang, Y.-P.; Chen, Y.-H.; Lin, C.-T.; Li, W.-S.; Wu, H.-C. Liposomal paclitaxel induces fewer hematopoietic and cardiovascular complications than bioequivalent doses of Taxol. Int. J. Oncol. 2018, 53, 1105–1117. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, R. Tarascon Pocket Phramacopoeia, Deluxe Lab-Coat Edition; Jones & Bartlett Learning: Burlington, MA, USA, 2015; Volume 54, ISBN 9781284057560. [Google Scholar]
- Heppler, L.N.; Walker, S.R.; Attarha, S.; Page, B.D.; Frank, D.A. Pyrimethamine inhibits STAT3 transcriptional activity via dihydrofolate reductase. In Proceedings of the AACR Annual Meeting, Atlanta, GA, USA, 29 March–3 April 2019. AM2019-387. [Google Scholar] [CrossRef]
- Song, J.S.; Rho, H.J.; Park, J.S.; Kim, M.S.; Lee, B.H.; Seo, J.W.; Jeon, D.J.; Cheon, H.G.; Ahn, S.H.; Kwon, K.; et al. Preclinical pharmacokinetics of PDE-310, a novel PDE4 inhibitor. Drug Metab. Pharmacokinet. 2011, 26, 192–200. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| Cell Line | DX2 Level | GI50 (µM) 1 | |
|---|---|---|---|
| Pyrimethamine | Taxol | ||
| H460 | high | 0.01 | 0.02 |
| A549 HCC-1359 HCC-366 | moderate | 0.8 14.3 22.3 | 0.03 0.08 0.05 |
| WI-26 H2087 | low | >100 >100 | 0.04 0.03 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.G.; Park, C.M.; Huddar, S.; Lim, S.; Kim, S.; Lee, S. Anticancer Activity of Pyrimethamine via Ubiquitin Mediated Degradation of AIMP2-DX2. Molecules 2020, 25, 2763. https://doi.org/10.3390/molecules25122763
Kim DG, Park CM, Huddar S, Lim S, Kim S, Lee S. Anticancer Activity of Pyrimethamine via Ubiquitin Mediated Degradation of AIMP2-DX2. Molecules. 2020; 25(12):2763. https://doi.org/10.3390/molecules25122763
Chicago/Turabian StyleKim, Dae Gyu, Chul Min Park, Srigouri Huddar, Semi Lim, Sunghoon Kim, and Sunkyung Lee. 2020. "Anticancer Activity of Pyrimethamine via Ubiquitin Mediated Degradation of AIMP2-DX2" Molecules 25, no. 12: 2763. https://doi.org/10.3390/molecules25122763
APA StyleKim, D. G., Park, C. M., Huddar, S., Lim, S., Kim, S., & Lee, S. (2020). Anticancer Activity of Pyrimethamine via Ubiquitin Mediated Degradation of AIMP2-DX2. Molecules, 25(12), 2763. https://doi.org/10.3390/molecules25122763