Induction of Redox-Mediated Cell Death in ER-Positive and ER-Negative Breast Cancer Cells by a Copper(II)-Phenolate Complex: An In Vitro and In Silico Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Copper Complexes
2.2. Cell Culture
2.3. Cytotoxicity Assay
2.4. AO & EB Fluorescent Probe for Assessment of Cell Death
2.5. Annexin V-Cy3 Apoptosis Assay
2.6. Measurement of ROS
2.7. Assay of Mitochondrial Trans-Membrane Potential (JC-1 Staining)
2.8. Alkaline Single-Cell Gel Electrophoresis (Comet) Assay
2.9. Cell Cycle Analysis
2.10. Western Blot Analysis
2.11. ADMET and Molecular Docking Studies
2.12. Statistical Analysis
3. Results
3.1. Cytotoxic Potential of the Complex as Revealed in MTT Assay
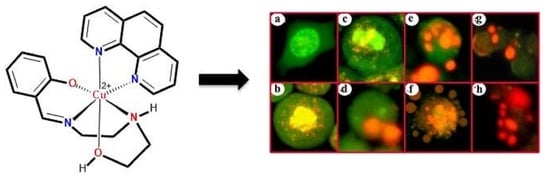
3.2. Indications of Apoptosis and Necrosis as Revealed in AO-EB Staining
3.3. Indications of Early Apoptotic Changes as Revealed in Annexin V-Cy3 and 6-CFDA Staining
3.4. Changes in ROS Level
3.5. Change in the Mitochondrial Membrane Potential as Revealed in JC-1 Staining
3.6. DNA Damage as Revealed in Comet Assay
3.7. Change in Cell Cycle Progression as Revealed in Flow Cytometric Analysis
3.8. Change in Molecular Indicators of Apoptosis
3.9. ADMET and Molecular Docking Studies
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy—An update from drug design perspective. Drug Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frezza, M.; Hindo, S.; Chen, D.; Davenport, A.; Schmitt, S.; Tomco, D.; Dou, Q.P. Novel metals and metal complexes as platforms for cancer therapy. Curr. Pharm. Des. 2010, 16, 1813–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Dilruba, S.; Kalayda, G.V. Platinum-based drugs: Past, present and future. Cancer Chemother. Pharmacol. 2016, 77, 1103–1124. [Google Scholar] [CrossRef] [PubMed]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef] [PubMed]
- Manohar, S.; Leung, N. Cisplatin nephrotoxicity: A review of the literature. J. Nephrol. 2018, 31, 15–25. [Google Scholar] [CrossRef]
- Nardon, C.; Boscutti, G.; Fregona, D. Beyond platinums: Gold complexes as anticancer agents. Anticancer Res. 2014, 34, 487–492. [Google Scholar]
- Ott, I.; Gust, R. Non platinum metal complexes as anti-cancer drugs. Arch. Pharm. 2007, 340, 117–126. [Google Scholar] [CrossRef]
- Zhao, G.; Lin, H. Metal complexes with aromatic N-containing ligands as potential agents in cancer treatment. AntiCancer Agents Med. Chem. 2005, 5, 137–147. [Google Scholar] [CrossRef]
- Kuduk-Jaworska, J.; Puszko, A.; Kubiak, M.; Pelczynska, M. Synthesis, structural, physico-chemical and biological properties of new palladium(II) complexes with 2,6-dimethyl-4-nitropyridine. J. Inorg. Biochem. 2004, 98, 1447–1456. [Google Scholar] [CrossRef]
- Hu, J.; Liao, C.; Mao, R.; Zhang, J.; Zhao, J.; Gu, Z. DNA interactions and in vitro anticancer evaluations of pyridine-benzimidazole-based Cu complexes. Med. Chem. Comm. 2018, 9, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.F.; Orvig, C.; Liang, H. Multi-target metal-based anticancer agents. Curr. Top. Med. Chem. 2017, 17, 3131–3145. [Google Scholar] [CrossRef] [PubMed]
- Nobili, S.; Mini, E.; Landini, I.; Gabbiani, C.; Casini, A.; Messori, L. Gold compounds as anticancer agents: Chemistry, cellular pharmacology, and preclinical studies. Med. Res. Rev. 2010, 30, 550–580. [Google Scholar] [CrossRef] [PubMed]
- Prosser, K.E.; Chang, S.W.; Saraci, F.; Le, P.H.; Walsby, C.J. Anticancer copper pyridine benzimidazole complexes: ROS generation, biomolecule interactions, and cytotoxicity. J. Inorg. Biochem. 2017, 167, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Wan, D.; Lai, S.H.; Yang, H.H.; Zhang, C.; Wang, X.Z.; Zeng, C.C.; Liu, Y.J. Design, synthesis and evaluation of anticancer activity of ruthenium (II) polypyridyl complexes. J. Inorg. Biochem. 2017, 173, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wu, Q.; Wang, C.; Tan, W.; Mei, W. Ruthenium(II) complexes as potential apoptosis inducers in chemotherapy. AntiCancer Agents Med. Chem. 2017, 17, 29–39. [Google Scholar]
- Li, W.; Jiang, G.B.; Yao, J.H.; Wang, X.Z.; Wang, J.; Han, B.J.; Xie, Y.Y.; Lin, G.J.; Huang, H.L.; Liu, Y.J. Ruthenium(II) complexes: DNA-binding, cytotoxicity, apoptosis, cellular localization, cell cycle arrest, reactive oxygen species, mitochondrial membrane potential and western blot analysis. J. Photochem. Photobiol. B 2014, 140, 94–104. [Google Scholar] [CrossRef]
- Xu, L.; Zhong, N.J.; Xie, Y.Y.; Huang, H.L.; Jiang, G.B.; Liu, Y.J. Synthesis, characterization, in vitro cytotoxicity, and apoptosis-inducing properties of ruthenium(II) complexes. PLoS ONE 2014, 9, e96082. [Google Scholar] [CrossRef]
- Misirlic-Dencic, S.; Poljarevic, J.; Isakovic, A.M.; Sabo, T.; Markovic, I.; Trajkovic, V. Current development of metal complexes with diamine ligands as potential anticancer agents. Curr. Med. Chem. 2020, 27, 380–410. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef]
- Shobha Devi, C.; Thulasiram, B.; Aerva, R.R.; Nagababu, P. Recent advances in copper intercalators as anticancer agents. J. Fluoresc. 2018, 28, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Portugal, J.; Barceló, F. Noncovalent binding to DNA: Still a target in developing anticancer agents. Curr. Med. Chem. 2016, 23, 4108–4134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef] [Green Version]
- Tapiero, H.; Townsend, D.M.; Tew, K.D. Trace elements in human physiology and pathology. Copper. Biomed. Pharmacother. 2003, 57, 386–398. [Google Scholar] [CrossRef]
- Sharma, M.; Ganeshpandian, M.; Majumder, M.; Tamilarasan, A.; Mukhopadhyay, R.; Islam, N.S.; Palaniandavar, M. Octahedral copper(II)-diimine complexes of triethylenetetramine: Effect of stereochemical fluxionality and ligand hydrophobicity on Cu(II)/Cu(I) redox, DNA binding and cleavage, cytotoxicity and apoptosis-inducing ability. Dalton Trans. 2020, 49, 8282–8297. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Shakthipriya, D.; Suresh, E.; Periasamy, V.S.; Akbarsha, M.A.; Palaniandavar, M. Ternary dinuclear copper(II) complexes of a hydroxybenzamide ligand with diimine coligands: The 5,6-dmp ligand enhances DNA binding and cleavage and induces apoptosis. Inorg. Chem. 2011, 50, 6458–6471. [Google Scholar] [CrossRef]
- Maciel, L.L.F.; de Freitas, W.R.; Bull, E.S.; Fernandes, C.; Horn, A.; de Aquino Almeida, J.C.; Kanashiro, M.M. In vitro and in vivo anti-proliferative activity and ultrastructure investigations of a copper(II) complex toward human lung cancer cell NCI-H460. J. Inorg. Biochem. 2020, 210, 111166. [Google Scholar] [CrossRef]
- Parsekar, S.U.; Singh, M.; Mishra, D.P.; Antharjanam, P.K.S.; Koley, A.P.; Kumar, M. Efficient hydrolytic cleavage of DNA and antiproliferative effect on human cancer cells by two dinuclear Cu(II) complexes containing a carbohydrazone ligand and 1,10-phenanthroline as a coligand. J. Biol. Inorg. Chem. 2019, 24, 343–363. [Google Scholar] [CrossRef]
- Polloni, L.; Seni Silva, A.C.; Teixeira, S.C.; Azevedo, F.V.P.V.; Zóia, M.A.P.; da Silva, M.S.; Lima, P.M.A.P.; Correia, L.I.V.; do Couto Almeida, J.; da Silva, C.V.; et al. Action of copper(II) complex with β-diketone and 1,10-phenanthroline (CBP-01) on sarcoma cells and biological effects under cell death. Biomed. Pharmacother. 2019, 112, 108586. [Google Scholar] [CrossRef]
- Haleel, A.; Mahendiran, D.; Veena, V.; Sakthivel, N.; Rahiman, A.K. Antioxidant, DNA interaction, VEGFR2 kinase, topoisomerase I and in vitro cytotoxic activities of heteroleptic copper(II) complexes of tetrazolo[1,5-a]pyrimidines and diimines. Mat. Sci. Eng. C. Mater. 2016, 68, 366–382. [Google Scholar] [CrossRef]
- Ng, C.H.; Chan, C.W.; Lai, J.W.; Ooi, I.H.; Chong, K.V.; Maah, M.J.; Seng, H.L. Enantiomeric pair of copper(II) polypyridyl-alanine complexes: Effect of chirality on their interaction with biomolecules. J. Inorg. Biochem. 2016, 160, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Xu, J.; Wang, Y.; Yu, H.; Yang, Y.; Liu, Y.; Ding, W.; Zhu, W.; Chen, R.; Ge, Z.; et al. Ternary copper(II) complexes with amino acid chains and heterocyclic bases: DNA binding, cytotoxic and cell apoptosis induction properties. J. Inorg. Biochem. 2015, 144, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Chen, R.; Yu, H.; Feng, Y.; Chen, J.; Lu, Q.; Xie, J.; Ding, W.; Ma, T. Antitumor effect of a copper (II) complex of a coumarin derivative and phenanthroline on lung adenocarcinoma cells and the mechanism of action. Mol. Med. Rep. 2014, 10, 2477–2482. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Chen, W.; Yin, Y.; Zheng, Z.; Zou, G. Probing the cell death signaling pathway of HepG2 cell line induced by copper-1,10-phenanthroline complex. Biometals 2014, 27, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zheng, C.; Zou, G.; Tao, D.; Gong, J. G(1)-phase specific apoptosis in liver carcinoma cell line induced by copper-1,10-phenanthroline. Int. J. Biochem. Cell. Biol. 2002, 34, 678–684. [Google Scholar] [CrossRef]
- Shi, X.; Fang, H.; Guo, Y.; Yuan, H.; Guo, Z.; Wang, X. Anticancer copper complex with nucleus, mitochondrion and cyclooxygenase-2 as multiple targets. J. Inorg. Biochem. 2019, 190, 38–44. [Google Scholar] [CrossRef]
- Slator, C.; Barron, N.; Howe, O.; Kellett, A. [Cu(o-phthalate)(phenanthroline)] exhibits unique superoxide-mediated NCI-60 chemotherapeutic action through genomic DNA damage and mitochondrial dysfunction. ACS Chem. Biol. 2016, 11, 159–171. [Google Scholar] [CrossRef]
- Gaál, A.; Orgován, G.; Mihucz, V.G.; Pape, I.; Ingerle, D.; Streli, C.; Szoboszlai, N. Metal transport capabilities of anticancer copper chelators. J. Trace Elem. Med. Biol. 2018, 47, 79–88. [Google Scholar] [CrossRef]
- Mukherjee, N.; Podder, S.; Banerjee, S.; Majumdar, S.; Nandi, D.; Chakravarty, A.R. Targeted photocytotoxicity by copper(II) complexes having vitamin B6 and photoactive acridine moieties. Eur. J. Med. Chem. 2016, 122, 497–509. [Google Scholar] [CrossRef]
- Baig, S.; Seevasant, I.; Mohamad, J.; Mukheem, A.; Huri, H.Z.; Kamarul, T. Potential of apoptotic pathway-targeted cancer therapeutic research: Where do we stand? Cell Death Discov. 2016, 7, e2058. [Google Scholar] [CrossRef] [Green Version]
- Fulda, S. Targeting apoptosis signaling pathways for anticancer therapy. Front. Oncol. 2011, 1, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jendrossek, V. The intrinsic apoptosis pathways as a target in anticancer therapy. Curr. Pharm. Biotechnol. 2012, 13, 1426–1438. [Google Scholar] [CrossRef]
- Pfeffer, C.M.; Singh, A.T.K. Apoptosis: A target for anticancer therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Karakhanova, S.; Hartwig, W.; D’Haese, J.G.; Philippov, P.P.; Werner, J.; Bazhin, A.V. Mitochondria and mitochondrial ROS in cancer: Novel targets for anticancer therapy. J. Cell. Physiol. 2016, 231, 2570–2581. [Google Scholar] [CrossRef] [PubMed]
- Rochford, G.; Molphy, Z.; Kavanagh, K.; McCann, M.; Devereux, M.; Kellett, A.; Howe, O. Cu(II) phenanthroline-phenazine complexes dysregulate mitochondrial function and stimulate apoptosis. Metallomics 2020, 12, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Masuri, S.; Cadoni, E.; Cabiddu, M.G.; Isaia, F.; Demuru, M.G.; Moráň, L.; Buček, D.; Vaňhara, P.; Havel, J.; Pivetta, T. The first copper(II) complex with 1,10-phenanthroline and salubrinal with interesting biochemical properties. Metallomics 2020, 12, 891–901. [Google Scholar] [CrossRef]
- Parton, M.; Krajewski, S.; Smith, I.; Krajewska, M.; Archer, C.; Naito, M.; Ahern, R.; Reed, J.; Dowsett, M. Coordinate expression of apoptosis-associated proteins in human breast cancer before and during chemotherapy. Clin. Cancer Res. 2002, 8, 2100–2108. [Google Scholar]
- Michel, L.L.; von Au, A.; Mavratzas, A.; Smetanay, K.; Schütz, F.; Schneeweiss, A. Immune checkpoint blockade in patients with triple-negative breast cancer. Target. Oncol. 2020, 15, 415–428. [Google Scholar] [CrossRef]
- Fedele, P.; Sanna, V.; Fancellu, A.; Cinieri, S. A clinical evaluation of treatments that target cell cycle machinery in breast cancer. Expert Opin. Pharmacother. 2019, 20, 2305–2315. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast cancer treatment: A review. J. Am. Med. Assoc. 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Rajendiran, V.; Karthik, R.; Palaniandavar, M.; Stoeckli-Evans, H.; Periasamy, V.S.; Akbarsha, M.A.; Srinag, B.S.; Krishnamurthy, H. Mixed-ligand copper(II)-phenolate complexes: Effect of coligand on enhanced DNA and protein binding, DNA cleavage, and anticancer activity. Inorg. Chem. 2007, 46, 8208–8221. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Spector, D.L.; Goldman, R.D.; Leinwand, L.A. Culture and Biochemical Analysis of Cells. Cell: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1998. [Google Scholar]
- Wang, H.; Joseph, J.A. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 1999, 27, 612–616. [Google Scholar] [CrossRef]
- Gopalakrishna, P.; Khar, A. Comet assay to measure DNA damage in apoptotic cells. J. Proteom. 1995, 30, 69–73. [Google Scholar] [CrossRef]
- Riccardi, C.; Nicoletti, I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006, 1, 1458–1461. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Proliferat. 2012, 45, 487–498. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Lowe, S.W.; Ruley, H.E.; Jacks, T.; Housman, D.E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 1993, 74, 957–967. [Google Scholar] [CrossRef]
- Kyzioł, A.; Cierniak, A.; Gubernator, J.; Markowski, A.; Jeżowska-Bojczuk, M.; Komarnicka, U.K. Copper(I) complexes with phosphine derived from sparfloxacin. Part III: Multifaceted cell death and preliminary study of liposomal formulation of selected copper(I) complexes. Dalton Trans. 2018, 47, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, R.; Ganeshpandian, M.; Bhuvanesh, N.S.P.; Palaniandavar, M.; Muruganantham, A.; Ghosh, S.K.; Riyasdeen, A.; Akbarsha, M.A. DNA and protein binding, double-strand DNA cleavage and cytotoxicity of mixed ligand copper(II) complexes of the antibacterial drug nalidixic acid. J. Inorg. Biochem. 2017, 174, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Rajendiran, V.; Palaniandavar, M.; Periasamy, V.S.; Srinag, B.S.; Krishnamurthy, H.; Akbarsha, M.A. Induction of cell death by ternary copper(II) complexes of L-tyrosine and diimines: Role of coligands on DNA binding and cleavage and anticancer activity. Inorg. Chem. 2009, 48, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Gandin, V.; Porchia, M.; Tisato, F.; Zanella, A.; Severin, E.; Dolmella, A.; Marzano, C. Novel mixed-ligand copper(I) complexes: Role of diimine ligands on cytotoxicity and genotoxicity. J. Med. Chem. 2013, 56, 7416–7430. [Google Scholar] [CrossRef]
- Molphy, Z.; Slator, C.; Chatgilialoglu, C.; Kellett, A. DNA oxidation profiles of copper phenanthrene chemical nucleases. Front. Chem. 2015, 3, 28. [Google Scholar] [CrossRef] [Green Version]
- Wong, D.Y.Q.; Lim, J.H.; Ang, W.H. Induction of targeted necrosis with HER2-targeted platinum(IV) anticancer prodrugs. Chem. Sci. 2015, 6, 3051–3056. [Google Scholar] [CrossRef] [Green Version]
- Gençkal, H.M.; Erkisa, M.; Alper, P.; Sahin, S.; Ulukaya, E.; Ari, F. Mixed ligand complexes of Co(II), Ni(II) and Cu(II) with quercetin and diimine ligands: Synthesis, characterization, anti-cancer and anti-oxidant activity. J. Biol. Inorg. Chem. 2020, 25, 161–177. [Google Scholar] [CrossRef]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef]
- Ng, C.H.; Kong, S.M.; Tiong, Y.L.; Maah, M.J.; Sukram, N.; Ahmad, M.; Khoo, A.S. Selective anticancer copper(II)-mixed ligand complexes: Targeting of ROS and proteasomes. Metallomics 2014, 6, 892–906. [Google Scholar] [CrossRef]
- Guo, W.J.; Ye, S.S.; Cao, N.; Huang, J.; Gao, J.; Chen, Q.Y. ROS-mediated autophagy was involved in cancer cell death induced by novel copper(II) complex. Exp. Toxicol. Pathol. 2010, 62, 577–582. [Google Scholar] [CrossRef]
- Saraste, A.; Pulkki, K. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc. Res. 2000, 45, 528–537. [Google Scholar] [CrossRef]
- Grasso, D.; Zampieri, L.X.; Capelôa, T.; Van de Velde, J.A.; Sonveaux, P. Mitochondria in cancer. Cell Stress 2020, 4, 114–146. [Google Scholar] [CrossRef] [PubMed]
- Heijink, A.M.; Krajewska, M.; van Vugt, M.A. The DNA damage response during mitosis. Mutat. Res. 2013, 750, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Pietenpol, J.A.; Stewart, Z.A. Cell cycle checkpoint signaling: Cell cycle arrest versus apoptosis. Toxicology 2002, 181–182, 475–481. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, Y. Targeting p53 for novel anticancer therapy. Transl. Oncol. 2010, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Dalla Via, L.; García-Argáez, A.N.; Martínez-Vázquez, M.; Grancara, S.; Martinis, P.; Toninello, A. Mitochondrial permeability transition as target of anticancer drugs. Curr. Pharm. Des. 2014, 20, 223–244. [Google Scholar]
- Morisaki, T.; Katano, M. Mitochondria-targeting therapeutic strategies for overcoming chemoresistance and progression of cancer. Curr. Med. Chem. 2003, 10, 2517–2521. [Google Scholar] [CrossRef]
- Merino, D.; Lok, S.W.; Visvader, J.E.; Lindeman, G.J. Targeting BCL-2 to enhance vulnerability to therapy in estrogen receptor-positive breast cancer. Oncogene 2016, 35, 1877–1887. [Google Scholar] [CrossRef]
- Cory, S.; Adams, J.M. Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer Cell 2005, 8, 5–6. [Google Scholar] [CrossRef] [Green Version]
- Bailey, S.T.; Shin, H.; Westerling, T.; Liu, X.S.; Brown, M. Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 18060–18065. [Google Scholar] [CrossRef] [Green Version]
- Tomicic, M.T.; Kaina, B. Topoisomerase degradation, DSB repair, p53 and IAPs in cancer cell resistance to camptothecin-like topoisomerase I inhibitors. Biochim. Biophys. Acta Rev. Cancer 2013, 1835, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Beretta, G.L.; Perego, P.; Zunino, F. Targeting topoisomerase I: Molecular mechanisms and cellular determinants of response to topoisomerase I inhibitors. Expert. Opin. Ther. Targets 2008, 12, 1243–1256. [Google Scholar] [CrossRef] [PubMed]
- Sordet, O.; Khan, Q.A.; Kohn, K.W.; Pommier, Y. Apoptosis induced by topoisomerase inhibitors. Anticancer Agents Med. Chem. 2003, 3, 271–290. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |












| Cell Lines | 24 h | 48 h | ||||
|---|---|---|---|---|---|---|
| IC50 | m | r | IC50 | m | r | |
| (µM) | (µM) | |||||
| MCF-7 | 1.6 ± 0.8 | 2.9 | 0.8 | 1.2 ± 0.8 | 2.8 | 0.8 |
| MDA-MB-231 | 1.9 ± 1.2 | 3.1 | 0.9 | 1.0 ± 0.9 | 2.2 | 0.9 |
| Complexes | Duration | Sub-G0 | SD | G0+G1 | SD | S | SD | G2+M | SD |
|---|---|---|---|---|---|---|---|---|---|
| MDA-MB-231 | |||||||||
| Control | 24 h | 5.05 | ±0.6 | 80.77 | ±0.29 | 4.47 | ±0 | 7.46 | ±0.3 |
| [Cu(tdp)phen]+ | 1.82 | ±0.2 | 59.84 | ±0.46 | 19.4 | ±0.1 | 14.9 | ±0.2 | |
| Control | 48 h | 5.38 | ±0.7 | 83.88 | ±1.09 | 3.31 | ±0.2 | 5.96 | ±0.4 |
| [Cu(tdp)phen]+ | 3.69 | ±0.3 | 53.59 | ±1.02 | 17.1 | ±0.2 | 20.9 | ±0.4 | |
| MCF-7 | |||||||||
| Control | 24 h | 2.74 | ±0.3 | 77.39 | ±0.72 | 6.74 | ±0.4 | 9.89 | ±0.5 |
| [Cu(tdp)phen]+ | 5.35 | ±1.8 | 53.28 | ±8.88 | 16.4 | ±2.5 | 17.4 | ±3.7 | |
| Control | 48 h | 2.41 | ±0.4 | 54.84 | ±3.4 | 22.2 | ±1.8 | 16.8 | ±1.8 |
| [Cu(tdp)phen]+ | 25.72 | ±4.5 | 21.63 | ±0.31 | 15.8 | ±1 | 28.3 | ±2.7 | |
| Descriptors | Score |
|---|---|
| ADMET AlogP98 (partition coefficient) | 3.588 |
| ADMET Aqueous Solubility | −7.391 |
| ADMET Absorption Level | 0 |
| ADMET PSA2D—(Fast polar surface area) | 31.909 |
| ADMET EXT PPB | 2.89411 |
| ADMET BBB | 0.45 |
| ADMET EXT CYP2D6 | −2.30385 |
| ADMET EXT Hepatotoxicity | −2.52683 |
| TOPKAT Ames mutagenicity | Non-Mutagen |
| TOPKAT Ames mutagenicity score | −2.33954 |
| TOPKAT Carcinogenicity in female rat | Non-Carcinogen |
| TOPKAT Carcinogenicity in male rat | Non-Carcinogen |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Periasamy, V.S.; Riyasdeen, A.; Rajendiran, V.; Palaniandavar, M.; Krishnamurthy, H.; Alshatwi, A.A.; Akbarsha, M.A. Induction of Redox-Mediated Cell Death in ER-Positive and ER-Negative Breast Cancer Cells by a Copper(II)-Phenolate Complex: An In Vitro and In Silico Study. Molecules 2020, 25, 4504. https://doi.org/10.3390/molecules25194504
Periasamy VS, Riyasdeen A, Rajendiran V, Palaniandavar M, Krishnamurthy H, Alshatwi AA, Akbarsha MA. Induction of Redox-Mediated Cell Death in ER-Positive and ER-Negative Breast Cancer Cells by a Copper(II)-Phenolate Complex: An In Vitro and In Silico Study. Molecules. 2020; 25(19):4504. https://doi.org/10.3390/molecules25194504
Chicago/Turabian StylePeriasamy, Vaiyapuri Subbarayan, Anvarbatcha Riyasdeen, Venugopal Rajendiran, Mallayan Palaniandavar, Hanumanthappa Krishnamurthy, Ali Abdullah Alshatwi, and Mohammad Abdulkader Akbarsha. 2020. "Induction of Redox-Mediated Cell Death in ER-Positive and ER-Negative Breast Cancer Cells by a Copper(II)-Phenolate Complex: An In Vitro and In Silico Study" Molecules 25, no. 19: 4504. https://doi.org/10.3390/molecules25194504
APA StylePeriasamy, V. S., Riyasdeen, A., Rajendiran, V., Palaniandavar, M., Krishnamurthy, H., Alshatwi, A. A., & Akbarsha, M. A. (2020). Induction of Redox-Mediated Cell Death in ER-Positive and ER-Negative Breast Cancer Cells by a Copper(II)-Phenolate Complex: An In Vitro and In Silico Study. Molecules, 25(19), 4504. https://doi.org/10.3390/molecules25194504