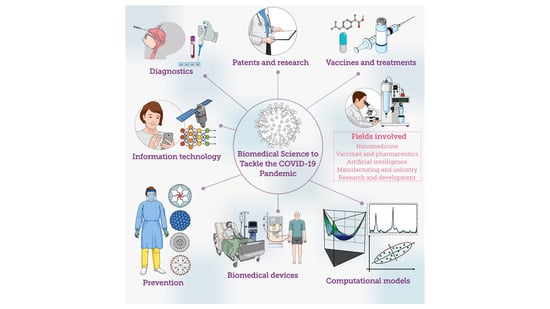
Biomedical Science to Tackle the COVID-19 Pandemic: Current Status and Future Perspectives
Abstract
:1. Introduction
2. Open Access to Designs and Patents
3. Biomedical Devices
4. Prevention of COVID-19 Infection Using Biomedical Science
4.1. Nosocomial Infections in Healthcare Facilities
4.2. Protective Personal Masks
4.3. Surface Decontamination
4.3.1. Silver Nanoparticles
4.3.2. Copper Oxide Nanoparticles
4.3.3. Titanium Oxide Nanoparticles
4.3.4. Other Oxide Nanoparticles
4.3.5. Graphene
5. Diagnostics and Biosensors for COVID-19
5.1. Non-Serological Methods
5.2. Serological Methods
5.2.1. Lateral Flow Methods
5.2.2. Surface Plasmon Resonance Techniques
5.2.3. Raman Spectroscopy Techniques
5.2.4. Colorimetric Techniques
5.2.5. Nanoparticle-Based Biosensing
6. Treatments for COVID-19
6.1. Computational Modeling for COVID-Related Structures
6.2. Artificial Intelligence
6.3. Small Molecule Therapeutics
6.4. Main Research Lines in Anti-COVID-19 Vaccines
7. Conclusions and Perspectives
Funding
Conflicts of Interest
Abbreviations
| ACE-2 | Angiotensin-Converting Enzyme 2 |
| AIDS | Acquired Immunodeficiency Syndrome |
| AI | Artificial Intelligence |
| CANARY | Cellular Analysis and Notification of Antigen Risks and Yields |
| CDC | US Center for Disease Control and Prevention |
| cDNA | Complementary DNA |
| CDR | Complementary-Determining Region |
| CISS | Chiral Induced Spin Selectivity |
| CL | Chemiluminescence |
| COVID-19 | Coronavirus Disease 2019 |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| ePPE | Electronic PPE |
| FDA | US Food and Drug Administration |
| GISAID | Global Initiative on Sharing All Influenza Data |
| GPS | Global Positioning System |
| GROMACS | Groningen Machine for Chemical Simulations |
| H1N1 | Influenza A virus subtype H1N1 |
| H9N2 | Influenza A virus subtype H9N2 |
| HIV | Human Immunodeficiency Virus |
| IgG | Immunoglobulin G |
| IgM | Immunoglobulin M |
| IoT | Internet of Things |
| ISO | International Organization for Standardization |
| LAMMPS | Large-scale Atomic/Molecular Massively Parallel Simulator |
| LAMP | Loop-Mediated Isothermal Amplification |
| LFIA | Lateral Flow Immunoassay |
| LOD | Limit of Detection |
| LSPR | Localized SPR |
| MD | Molecular Dynamics |
| MERS | Middle East Respiratory Syndrome |
| NIAID | National Institute of Allergy and Infectious Diseases |
| NMs | Nanomaterials |
| NPs | Nanoparticles |
| OSHA | US Occupational Safety and Health Administration |
| PCR | Polymerase Chain Reaction |
| PIGS | Prediction of Immunoglobulin Structure |
| HPV | Human papillomavirus |
| PPE | Personal Protective Equipment |
| QDs | Quantum Dots |
| qPCR | Quantitative PCR |
| ROS | Reactive Oxygen Species |
| RT-PCR | Reverse Transcription PCR |
| RT-qPCR | Reverse Transcription Quantitative PCR |
| SARS | Severe Acute Respiratory Syndrome |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| SERS | Surface Enhanced Raman Spectroscopy |
| SIV | Swine Influenza Virus |
| SPR | Surface Plasmon Resonance |
| ssDNA | single-stranded DNA |
| TMV | Tobacco Mosaic Virus |
| VLP | Virus-Like Particle Platform |
| VRC | Vaccine Research Center |
| WAM | Web Antibody Modeling |
| WHO | World Health Organization |
References
- Tu, Y.-F.; Chien, C.-S.; Yarmishyn, A.A.; Lin, Y.-T.; Luo, Y.-H.; Lai, W.-Y.; Yang, D.-M.; Chou, S.-J.; Yang, Y.-P.; Wang, M.-L.; et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allam, M.; Cai, S.; Ganesh, S.; Venkatesan, M.; Doodhwala, S.; Song, Z.; Hu, T.; Kumar, A.; Heit, J.; Coskun, A.F.; et al. COVID-19 Diagnostics, Tools, and Prevention. Diagnostics 2020, 10, 409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gharizadeh, B.; Lu, D.; Yue, J.; Yu, M.; Liu, Y.; Zhou, M. Navigating the Pandemic Response Life Cycle: Molecular Diagnostics and Immunoassays in the Context of COVID-19 Management. IEEE Rev. Biomed. Eng. 2020, 1. [Google Scholar] [CrossRef] [PubMed]
- Sivasankarapillai, V.S.; Pillai, A.M.; Rahdar, A.; Sobha, A.P.; Das, S.S.; Mitropoulos, A.C.; Mokarrar, M.H.; Kyzas, G.Z. On Facing the SARS-CoV-2 (COVID-19) with Combination of Nanomaterials and Medicine: Possible Strategies and First Challenges. Nanomaterials 2020, 10, 852. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, H.; Díaz-López, S.; Jarre, E.; Pacheco, H.; Méndez, W.; Zamora-Ledezma, E. NO2 levels after the COVID-19 lockdown in Ecuador: A trade-off between environment and human health. Urban Clim. 2020, 34, 100674. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkings University & Medicine Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/map.html (accessed on 3 August 2020).
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [Green Version]
- Demogines, A.; Farzan, M.; Sawyer, S.L. Evidence for ACE2-Utilizing Coronaviruses (CoVs) Related to Severe Acute Respiratory Syndrome CoV in Bats. J. Virol. 2012, 86, 6350–6353. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, B.D.; Mou, H.; Zhang, L.; Guo, Y.; He, W.; Ojha, A.; Parcells, M.S.; Luo, G.; Li, W.; Zhong, G.; et al. The SARS-CoV-2 Receptor-Binding Domain Elicits a Potent Neutralizing Response Without Antibody-Dependent Enhancement. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ngom, B.; Guo, Y.; Wang, X.; Bi, D. Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: A review. Anal. Bioanal. Chem. 2010, 397, 1113–1135. [Google Scholar] [CrossRef]
- Donnelly, C.A.; Malik, M.R.; Elkholy, A.; Cauchemez, S.; Van Kerkhove, M.D. Worldwide Reduction in MERS Cases and Deaths since 2016. Emerg. Infect. Dis. 2019, 25, 1758–1760. [Google Scholar] [CrossRef]
- Liu, C.Y.; Zhou, Q.A.; Li, Y.; Garner, L.V.; Watkins, S.P.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S.; et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Pero, J. Scientists and Tech Companies are Open-Sourcing Their Patents as Part of a Pledge to Help Expand Resources for Researchers Trying to Combat COVID-19 Pandemic. Available online: https://www.dailymail.co.uk/sciencetech/article-8206471/Scientists-tech-companies-open-source-patents-help-researchers-studying-COVID-19.html (accessed on 27 July 2020).
- Philippidis, A. Vanquishing the Virus: 160+ COVID-19 Drug and Vaccine Candidates in Development. Available online: https://www.genengnews.com/a-lists/vanquishing-the-virus-160-covid-19-drug-and-vaccine-candidates-in-development/ (accessed on 27 July 2020).
- Apuzzo, M.; David, D.K. Covid-19 Changed How the World Does Science, Together. Available online: https://www.nytimes.com/2020/04/01/world/europe/coronavirus-science-research-cooperation.html (accessed on 27 July 2020).
- Rourke, M.; Eccleston-Turner, M.; Phelan, A.; Gostin, L.O. Policy opportunities to enhance sharing for pandemic research. Science 2020, 368, 716–718. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FDA Emergency Use Authorization: Emergency Use Authorization (EUA) Information, and List of All Current EUAs. Available online: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization (accessed on 27 July 2020).
- Excelra Knowledge Solutions Pvt. Ltd. (EXCELRA). Available online: https://www.excelra.com/california-institute/ (accessed on 27 July 2020).
- Donato, P.M.; Hannah, K. AbbVie Drops Patent Rights for Kaletra Antiviral Treatment. Available online: https://www.ft.com/content/5a7a9658-6d1f-11ea-89df-41bea055720b (accessed on 27 July 2020).
- Pfizer and Biontech Announce Further Details on Collaboration to Accelerate Global Covid-19 Vaccine Development. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-announce-further-details-collaboration (accessed on 27 July 2020).
- Harvey Allchurch, M.; McCullough, G. Medicines Patent Pool and Unitaid Respond to Access Efforts for COVID-19 Treatments and Technologies. Available online: https://unitaid.org/news-blog/medicines-patent-pool-and-unitaid-respond-to-access-efforts-for-covid-19-treatments-and-technologies/#en (accessed on 27 July 2020).
- Courage, N.; Kovarik, R. Canadians Ramping up COVID-19 Collaborations and IP. Available online: https://www.bereskinparr.com/doc/canadians-ramping-up-covid-19-collaborations (accessed on 27 July 2020).
- Etherington, D. Medtronic is Sharing its Portable Ventilator Design Specifications and Code for Free to All. Available online: https://techcrunch.com/2020/03/30/medtronic-is-sharing-its-portable-ventilator-design-specifications-and-code-for-free-to-all/ (accessed on 28 July 2020).
- Peters, J. Volunteers Produce 3D-Printed Valves For Life-Saving Coronavirus Treatments. Available online: https://www.theverge.com/2020/3/17/21184308/coronavirus-italy-medical-3d-print-valves-treatments (accessed on 28 July 2020).
- Benchaita, S. IBM Tops U.S. Patent List for 2019. Available online: https://newsroom.ibm.com/2020-01-14-IBM-Tops-U-S-Patent-List-for-2019 (accessed on 28 July 2020).
- Ringes, M. IBM Offering Free Access to Patent Portfolio to Combat COVID-19. Available online: https://www.ibm.com/blogs/research/2020/04/ibm-patent-portfolio-access-combat-covid-19/ (accessed on 28 July 2020).
- Cohen, D. Tech Giants Join the Open Covid Pledge to Make Their Patents Available at No Cost. Available online: https://www.adweek.com/digital/tech-giants-join-the-open-covid-pledge-to-make-their-patents-available-at-no-cost/ (accessed on 28 July 2020).
- Dearborn, M. Ford Works with 3M, GE, UAW to Speed Production of Respirators for Healthcare Workers, Ventilators for Coronavirus Patients. Available online: https://media.ford.com/content/fordmedia/fna/us/en/news/2020/03/24/ford-3m-ge-uaw-respirators-ventilators.html (accessed on 28 July 2020).
- Santavicca, G. Intellectual Property Resilience in the Era of Covid-19. Available online: http://www.medialaws.eu/intellectual-property-resilience-in-the-era-of-covid-19/ (accessed on 28 July 2020).
- Dargaville, T.; Spann, K.; Celina, M. Opinion to address the personal protective equipment shortage in the global community during the COVID-19 outbreak. Polym. Degrad. Stab. 2020, 176, 109162. [Google Scholar] [CrossRef] [PubMed]
- Jessop, Z.M.; Dobbs, T.D.; Ali, S.R.; Combellack, E.; Clancy, R.; Ibrahim, N.; Jovic, T.H.; Kaur, A.J.; Nijran, A.; O’Neill, T.B.; et al. Personal Protective Equipment (PPE) for Surgeons during COVID-19 Pandemic: A Systematic Review of Availability, Usage, and Rationing. Br. J. Surg. 2020. [Google Scholar] [CrossRef]
- Harvard University Graduate School of Design GSD Begins Patient Isolation Hood (PIH) Design and Fabrication Alongside Ongoing PPE Efforts. Available online: https://www.gsd.harvard.edu/2020/04/gsd-begins-patient-isolation-hood-pih-design-and-fabrication-alongside-ongoing-ppe-efforts/ (accessed on 3 August 2020).
- Turer, R.W.; Jones, I.; Rosenbloom, S.T.; Slovis, C.; Ward, M.J. Electronic personal protective equipment: A strategy to protect emergency department providers in the age of COVID-19. J. Am. Med. Inf. Assoc. 2020, 27, 967–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- About the Hemolung RAS. Available online: https://www.alung.com/hemolung-ras-us/ (accessed on 30 July 2020).
- Ravenscroft, T. Weston Williamson + Partners Envisions Social-Distancing Office. Available online: https://www.dezeen.com/2020/05/14/weston-williamson-social-distancing-office/?li_source=LI&li_medium=bottom_block_1 (accessed on 3 August 2020).
- Abeler, J.; Bäcker, M.; Buermeyer, U.; Zillessen, H. COVID-19 Contact Tracing and Data Protection Can Go Together. JMIR mHealth uHealth 2020, 8, e19359. [Google Scholar] [CrossRef] [Green Version]
- Vaishya, R.; Javaid, M.; Khan, I.H.; Haleem, A. Artificial Intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 337–339. [Google Scholar] [CrossRef]
- Pagliacolo, E.; Pavka, E. Design Takes Action: Innovations for the COVID-19 Crisis. Available online: https://www.azuremagazine.com/article/design-innovations-covid-19-ppe/ (accessed on 3 August 2020).
- The Pneumask Project. Available online: https://www.pneumask.org (accessed on 3 August 2020).
- Maker Mask: 3D Printable Respirator quality Masks. Available online: https://www.makermask.com/ (accessed on 3 August 2020).
- Duke University: Technologies & Start-Ups. Available online: https://olv.duke.edu/covid-19/technologies-and-startups/#surgical-hoods (accessed on 3 August 2020).
- MIT News. MIT Initiates Mass Manufacture of Disposable Face Shields for Covid-19 Response. Available online: https://news.mit.edu/2020/face-shield-ppe-manufacture-covid-19-0331 (accessed on 3 August 2020).
- National University of Singapore News. On-Site COVID-19 Test Results in One Hour. Available online: https://news.nus.edu.sg/research/portable-point-care-test-covid-19-diagnosis (accessed on 3 August 2020).
- GLASSAFE News & Events. Available online: http://aviointeriors.it/2020/press/glassafe/ (accessed on 3 August 2020).
- MIT Emergency Ventilator, Design Toolbox. Available online: https://emergency-vent.mit.edu/ (accessed on 3 August 2020).
- Georgia Tech News Center. Simple, Low-cost Ventilator Builds on Available Resuscitation Bags. Available online: https://news.gatech.edu/2020/04/06/simple-low-cost-ventilator-builds-available-resuscitation-bags (accessed on 3 August 2020).
- NASA. NASA Develops COVID-19 Prototype Ventilator in 37 Days. Available online: https://www.nasa.gov/feature/jpl/nasa-develops-covid-19-prototype-ventilator-in-37-days (accessed on 3 August 2020).
- Child, D. Evening Standard: Harvard and MIT Researchers Race to Develop Face Mask That Lights up When It Detects Coronavirus. Available online: https://www.standard.co.uk/news/world/harvard-mit-researchers-developing-flashing-coronavirus-face-mask-a4441441.html (accessed on 3 August 2020).
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef]
- WHO. Infection Prevention and Control during Health Care When COVID-19 Is Suspected or Confirmed; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Del Rio, C.; Malani, P.N. COVID-19—New Insights on a Rapidly Changing Epidemic. JAMA 2020, 323, 1339. [Google Scholar] [CrossRef] [Green Version]
- WHO. Coronavirus Disease 2019 (COVID-19) Situation Report–72; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Chan, K.H.; Peiris, J.S.M.; Lam, S.Y.; Poon, L.L.M.; Yuen, K.Y.; Seto, W.H. The Effects of Temperature and Relative Humidity on the Viability of the SARS Coronavirus. Adv. Virol. 2011, 2011, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Casanova, L.M.; Jeon, S.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Effects of Air Temperature and Relative Humidity on Coronavirus Survival on Surfaces. Appl. Environ. Microbiol. 2010, 76, 2712–2717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of surrogate coronaviruses in water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef] [PubMed]
- CDC. Interim Infection Prevention and Control Recommendations for Patients with Suspected Confirmed Coronavirus Disease 2019 (COVID019): Healthcare Settings; CDC: Atlanta, GA, USA, 2020. [Google Scholar]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhems, P.; Saadatian-Elahi, M.; Chuzeville, M.; Marion, E.; Favrelle, L.; Hilliquin, D.; Martin-Gaujard, G.; Gourmelon, R.; Noaillon, M.; Khanafer, N. Rapid nosocomial spread of SARS-CoV-2 in a French geriatric unit. Infect. Control. Hosp. Epidemiol. 2020, 41, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-C.; Huang, L.-M.; Chan, C.-C.; Su, C.-P.; Chang, S.-C.; Chang, Y.-Y.; Chen, M.-L.; Hung, C.-C.; Chen, W.-J.; Lin, F.-Y.; et al. SARS in Hospital Emergency Room. Emerg. Infect. Dis. 2004, 10, 782–788. [Google Scholar] [CrossRef]
- Ong, S.W.X.; Tan, Y.K.; Chia, P.Y.; Lee, T.H.; Ng, O.T.; Wong, M.S.Y.; Marimuthu, K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA 2020, 323, 1610. [Google Scholar] [CrossRef] [Green Version]
- Bridier, A.; Briandet, R.; Thomas, V.; Dubois-Brissonnet, F. Resistance of bacterial biofilms to disinfectants: A review. Biofouling 2011, 27, 1017–1032. [Google Scholar] [CrossRef]
- Köck, R.; Harlizius, J.; Bressan, N.; Laerberg, R.; Wieler, L.H.; Witte, W.; Deurenberg, R.H.; Voss, A.; Becker, K.; Friedrich, A.W. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among pigs on German farms and import of livestock-related MRSA into hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1375–1382. [Google Scholar] [CrossRef] [Green Version]
- Webb, G.F.; D’Agata, E.M.C.; Magal, P.; Ruan, S. A model of antibiotic-resistant bacterial epidemics in hospitals. Proc. Natl. Acad. Sci. USA 2005, 102, 13343–13348. [Google Scholar] [CrossRef] [Green Version]
- Lara, H.H.; Ayala-Núñez, N.V.; Ixtepan-Turrent, L.; Rodriguez-Padilla, C. Mode of antiviral action of silver nanoparticles against HIV-1. J. Nanobiotechnol. 2010, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Lara, H.H.; Ixtepan-Turrent, L.; Garza-Treviño, E.N.; Rodriguez-Padilla, C. PVP-coated silver nanoparticles block the transmission of cell-free and cell-associated HIV-1 in human cervical culture. J. Nanobiotechnol. 2010, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimori, Y.; Sato, T.; Hayata, T.; Nagao, T.; Nakayama, M.; Nakayama, T.; Sugamata, R.; Suzuki, K. Novel Antiviral Characteristics of Nanosized Copper(I) Iodide Particles Showing Inactivation Activity against 2009 Pandemic H1N1 Influenza Virus. Appl. Environ. Microbiol. 2011, 78, 951–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borkow, G.; Gabbay, J. Putting copper into action: Copper-impregnated products with potent biocidal activities. FASEB J. 2004, 18, 1728–1730. [Google Scholar] [CrossRef]
- Borkow, G.; Zhou, S.S.; Page, T.; Gabbay, J. A Novel Anti-Influenza Copper Oxide Containing Respiratory Face Mask. PLoS ONE 2010, 5, e11295. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.G.; Walls, R.M. Supporting the Health Care Workforce During the COVID-19 Global Epidemic. JAMA 2020, 323, 1439. [Google Scholar] [CrossRef] [Green Version]
- Holland, M.; Zaloga, D.J.; Friderici, C.S. COVID-19 Personal Protective Equipment (PPE) for the emergency physician. Vis. J. Emerg. Med. 2020, 19, 100740. [Google Scholar] [CrossRef]
- Aitken, C.; Jeffries, D.J. Nosocomial Spread of Viral Disease. Clin. Microbiol. Rev. 2001, 14, 528–546. [Google Scholar] [CrossRef] [Green Version]
- Luong-Nguyen, M.; Hermand, H.; Abdalla, S.; Cabrit, N.; Hobeika, C.; Brouquet, A.; Goéré, D.; Sauvanet, A. Nosocomial infection with SARS-Cov-2 within Departments of Digestive Surgery. J. Visc. Surg. 2020, 157, S13–S18. [Google Scholar] [CrossRef]
- Bellizzi, S.; Napodano, C.M.P.; Salaris, P.; Pichierri, G.; Sotgiu, G. Regional variation in trajectories of healthcare worker infections during the COVID-19 pandemic in Italy. Infect. Control. Hosp. Epidemiol. 2020, 1–3. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, M.; Liu, F. Reasons for healthcare workers becoming infected with novel coronavirus disease 2019 (COVID-19) in China. J. Hosp. Infect. 2020, 105, 100–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CDC. Non Occupational Uses of Respiratory Protection—What Public Health Organizations and Users Need to Know. 2018. Available online: https://blogs.cdc.gov/niosh-science-blog/2018/01/04/respirators-public-use (accessed on 27 July 2020).
- Tellier, R.; Li, Y.; Cowling, B.J.; Tang, J.W. Recognition of aerosol transmission of infectious agents: A commentary. BMC Infect. Dis. 2019, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.Y.; Weng, W.G.; Huang, Q.Y. Characterizations of particle size distribution of the droplets exhaled by sneeze. J. R. Soc. Interface 2013, 10, 20130560. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lee, G.W.-M.; Chen, C.-M.; Lee, C.-P.; Yu, K.-P. The Size and Concentration of Droplets Generated by Coughing in Human Subjects. J. Aerosol Med. 2007, 20, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- WHO. Updated WHO Recommendations for International Traffic in Relation to COVID-19 Outbreak. 2020. Available online: https://www.who.int/news-room/articlesdetail/updated-who-recommendations-for-internationaltraffic-in-relation-to-covid-19-outbreak (accessed on 14 April 2020).
- WHO. Management of Ill Travellers at Points of Entry (International Airports, Seaports, and Ground Crossings) in the Context of COVID-Interim Guidance. Available online: https://apps.who.int/iris/bitstream/handle/10665/331512/WHO-2019-nCoV-POEmgmt-2020.2-eng.pdf?sequence=1&isAllowed=y (accessed on 14 April 2020).
- CDC. Returning from International Travel; CDC: Atlanta, GA, USA, 2020. [Google Scholar]
- Bourouiba, L.; Dehandschoewercker, E.; Bush, J.W.M. Violent expiratory events: On coughing and sneezing. J. Fluid Mech. 2014, 745, 537–563. [Google Scholar] [CrossRef]
- Bourouiba, L. Turbulent Gas Clouds and Respiratory Pathogen Emissions. JAMA 2020. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Munster, V.J. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Eurosurveillance 2013, 18, 20590. [Google Scholar] [CrossRef] [Green Version]
- CDC. How Coronavirus Spreads. Available online: https://www.cdc.gov/coronavirus/2019-ncov/faq.html (accessed on 21 August 2020).
- CDC Weekly, C. The Epidemiological Characteristics of an Outbreak of 2019 Novel Coronavirus Diseases (COVID-19)—China. China CDC Wkly. 2020, 2, 113–122. [Google Scholar] [CrossRef]
- Mackay, I.M.; Arden, K. MERS coronavirus: Diagnostics, epidemiology and transmission. Virol. J. 2015, 12, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Memish, Z.A.; Al-Tawfiq, J.A. Middle East respiratory syndrome coronavirus: Epidemiology and disease control measures. Infect. Drug Resist. 2014, 7, 281–287. [Google Scholar] [CrossRef] [Green Version]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nat. Cell Biol. 2012, 486, 420–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasickova, P.; Pavlik, I.; Verani, M.; Carducci, A. Issues Concerning Survival of Viruses on Surfaces. Food Environ. Virol. 2010, 2, 24–34. [Google Scholar] [CrossRef]
- Seah, I.; Su, X.; Lingam, G. Revisiting the dangers of the coronavirus in the ophthalmology practice. Eye 2020, 34, 1155–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morawska, L. Droplet fate in indoor environments, or can we prevent the spread of infection? Indoor Air 2006, 16, 335–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaman, J.; Kohn, M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc. Natl. Acad. Sci. USA 2009, 106, 3243–3248. [Google Scholar] [CrossRef] [Green Version]
- Lowen, A.C.; Mubareka, S.; Steel, J.; Palese, P. Influenza Virus Transmission Is Dependent on Relative Humidity and Temperature. PLoS Pathog. 2007, 3, e151–e156. [Google Scholar] [CrossRef]
- Warnes, S.L.; Little, Z.R.; Keevil, C. Human Coronavirus 229E Remains Infectious on Common Touch Surface Materials. mBio 2015, 6, 01697–01715. [Google Scholar] [CrossRef] [Green Version]
- Rabenau, H.; Cinatl, J.; Morgenstern, B.; Bauer, G.; Preiser, W.; Doerr, H.W. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2004, 194, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Sizun, J.; Yu, M.; Talbot, P. Survival of human coronaviruses 229E and OC43 in suspension and after drying onsurfaces: A possible source ofhospital-acquired infections. J. Hosp. Infect. 2000, 46, 55–60. [Google Scholar] [CrossRef]
- Song, K.; Lee, J.; Choi, S.-O.; Kim, J. Interaction of Surface Energy Components between Solid and Liquid on Wettability, and Its Application to Textile Anti-Wetting Finish. Polymers 2019, 11, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chunglok, A.; Muensit, N.; Daengngam, C. Extreme Wetting-Resistant Multiscale Nano-/Microstructured Surfaces for Viscoelastic Liquid Repellence. J. Nanomater. 2016, 2016, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Deokar, A.R.; Nagvenkar, A.P.; Kalt, I.; Shani, L.; Yeshurun, Y.; Gedanken, A.; Sarid, R. Graphene-Based “Hot Plate” for the Capture and Destruction of the Herpes Simplex Virus Type 1. Bioconjug. Chem. 2017, 28, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-N.; Hsueh, Y.-H.; Hsieh, C.-T.; Tzou, D.-Y.; Chang, H.-T. Antiviral Activity of Graphene–Silver Nanocomposites against Non-Enveloped and Enveloped Viruses. Int. J. Environ. Res. Public Health 2016, 13, 430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, R.; Hara, M.; Ishiguro, H.; Yao, Y.; Ochiai, T.; Nakata, K.; Murakami, T.; Kajioka, J.; Sunada, K.; Hashimoto, K.; et al. Broad Spectrum Microbicidal Activity of Photocatalysis by TiO2. Catalysts 2013, 3, 310–323. [Google Scholar] [CrossRef] [Green Version]
- Akhavan, O.; Choobtashani, M.; Ghaderi, E. Protein Degradation and RNA Efflux of Viruses Photocatalyzed by Graphene–Tungsten Oxide Composite Under Visible Light Irradiation. J. Phys. Chem. C 2012, 116, 9653–9659. [Google Scholar] [CrossRef]
- Botequim, D.; Maia, J.; Ferreira, L.; Lopes, L.M.F.; Simões, P.N.; Ilharco, L.M.; Ferreira, L. Nanoparticles and Surfaces Presenting Antifungal, Antibacterial and Antiviral Properties. Langmuir 2012, 28, 7646–7656. [Google Scholar] [CrossRef]
- Nakano, R.; Ishiguro, H.; Yao, Y.; Kajioka, J.; Fujishima, A.; Sunada, K.; Minoshima, M.; Hashimoto, K.; Kubota, Y. Photocatalytic inactivation of influenza virus by titanium dioxide thin film. Photochem. Photobiol. Sci. 2012, 11, 1293–1298. [Google Scholar] [CrossRef]
- Mazurkova, N.A.; Spitsyna, Y.E.; Shikina, N.V.; Ismagilov, Z.R.; Zagrebel’Nyi, S.N.; Ryabchikova, E.I. Interaction of titanium dioxide nanoparticles with influenza virus. Nanotechnol. Russ. 2010, 5, 417–420. [Google Scholar] [CrossRef]
- Elechiguerra, J.L.; Burt, J.L.; Morones, J.R.; Camacho-Bragado, A.; Gao, X.; Lara, H.H.; Jose-Yacaman, M. Interaction of silver nanoparticles with HIV-1. J. Nanobiotechnol. 2005, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Xu, J.; Miao, D.; Peng, L.; Debeli, D.K.; Zhu, P. Water-repellency, ultraviolet protection and infrared emissivity properties of AZO film on polyester fabric. Ceram. Int. 2017, 43, 2424–2430. [Google Scholar] [CrossRef]
- Wahab, J.A.; Xu, G.; Lee, H.; Nam, P.D.; Wei, K.; Kim, S.H.; Kim, I.S. Fabrication of silk fibroin/eggshell nanofiber membranes for facemasks. Fibers Polym. 2016, 17, 1776–1781. [Google Scholar] [CrossRef]
- Warnes, S.L.; Keevil, C.W. Inactivation of Norovirus on Dry Copper Alloy Surfaces. PLoS ONE 2013, 8, e75017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunada, K.; Minoshima, M.; Hashimoto, K. Highly efficient antiviral and antibacterial activities of solid-state cuprous compounds. J. Hazard. Mater. 2012, 265–270. [Google Scholar] [CrossRef]
- Lazary, A.; Weinberg, I.; Vatine, J.-J.; Jefidoff, A.; Bardenstein, R.; Borkow, G.; Ohana, N. Reduction of healthcare-associated infections in a long-term care brain injury ward by replacing regular linens with biocidal copper oxide impregnated linens. Int. J. Infect. Dis. 2014, 24, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Burrer, S.L.; De Perio, M.A.; Hughes, M.M.; Kuhar, D.T.; Luckhaupt, S.E.; McDaniel, C.J.; Porter, R.M.; Silk, B.; Stuckey, M.J.; Walters, M. Characteristics of Health Care Personnel with COVID-19—United States, February 12–April 9, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 477–481. [Google Scholar] [CrossRef] [Green Version]
- Paxton, N.C.; Forrestal, D.P.; Desselle, M.; Kirrane, M.; Bugden, S.; Sullivan, C.; Powell, S.K.; Woodruff, M.A. N95 Respiratory Masks for COVID-19: A Review of the Literature to Inform Local Responses to Global Shortages. Available online: https://research.qut.edu.au/biofabrication/wp-content/uploads/sites/62/2020/04/N95_COVID-19_LiteratureReview_2020_Submission.pdf (accessed on 21 August 2020).
- Radonovich, L.J.; Simberkoff, M.S.; Bessesen, M.T.; Brown, A.C.; Cummings, D.A.T.; Gaydos, C.A.; Los, J.G.; Krosche, A.E.; Gibert, C.L.; Gorse, G.J.; et al. N95 Respirators vs. Medical Masks for Preventing Influenza Among Health Care Personnel. JAMA 2019, 322, 824–833. [Google Scholar] [CrossRef] [Green Version]
- Eninger, R.M.; Honda, T.; Adhikari, A.; Heinonen-Tanski, H.; Reponen, T.; Grinshpun, S.A. Filter Performance of N99 and N95 Facepiece Respirators Against Viruses and Ultrafine Particles. Ann. Occup. Hyg. 2008, 52, 385–396. [Google Scholar] [CrossRef]
- Zhong, H.; Zhu, Z.; Lin, J.; Cheung, C.F.; Lu, V.L.; Yan, F.; Chan, C.-Y.; Li, G. Reusable and Recyclable Graphene Masks with Outstanding Superhydrophobic and Photothermal Performances. ACS Nano 2020, 14, 6213–6221. [Google Scholar] [CrossRef]
- Tian, X.; Verho, T.; Ras, R.H.A. Moving superhydrophobic surfaces toward real-world applications. Science 2016, 352, 142–143. [Google Scholar] [CrossRef]
- Blecher, K.; Nasir, A.; Friedman, A. The growing role of nanotechnology in combating infectious disease. Virulence 2011, 2, 395–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narvaez-Muñoz, C.P.; Carrion-Matamoros, L.M.; Vizuete, K.; Debut, A.; Arroyo, C.R.; Guerrero, V.; Almeida-Naranjo, C.E.; Morales-Florez, V.; Mowbray, D.J.; Zamora-Ledezma, C. Tailoring Organic–Organic Poly(vinylpyrrolidone) Microparticles and Fibers with Multiwalled Carbon Nanotubes for Reinforced Composites. ACS Appl. Nano Mater. 2019, 2, 4302–4312. [Google Scholar] [CrossRef]
- Otter, J.; Donskey, C.; Yezli, S.; Douthwaite, S.; Goldenberg, S.; Weber, D. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: The possible role of dry surface contamination. J. Hosp. Infect. 2016, 92, 235–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firquet, S.; Beaujard, S.; Lobert, P.-E.; Sane, F.; Caloone, D.; Izard, D.; Hober, D. Survival of Enveloped and Non-Enveloped Viruses on Inanimate Surfaces. Microbes Environ. 2015, 30, 140–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharjee, S.; Joshi, R.; Chughtai, A.A.; MacIntyre, C.R. Graphene Modified Multifunctional Personal Protective Clothing. Adv. Mater. Interfaces 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Palza, H.; Núñez, M.; Bastías, R.; Delgado, K. In situ antimicrobial behavior of materials with copper-based additives in a hospital environment. Int. J. Antimicrob. Agents 2018, 51, 912–917. [Google Scholar] [CrossRef]
- Kumar, S.V.; Bafana, A.P.; Pawar, P.; Rahman, A.; Dahoumane, S.A.; Jeffryes, C.S. High conversion synthesis of <10 nm starch-stabilized silver nanoparticles using microwave technology. Sci. Rep. 2018, 8, 5106. [Google Scholar] [CrossRef] [Green Version]
- Castro-Mayorga, J.L.; Randazzo, W.; Fabra, M.J.; Lagaron, J.; Aznar, R.; Sanchez, G. Antiviral properties of silver nanoparticles against norovirus surrogates and their efficacy in coated polyhydroxyalkanoates systems. LWT Food. Sci. Technol. 2017, 79, 503–510. [Google Scholar] [CrossRef] [Green Version]
- Perreault, F.; De Faria, A.F.; Nejati, S.; Elimelech, M. Antimicrobial Properties of Graphene Oxide Nanosheets: Why Size Matters. ACS Nano 2015, 9, 7226–7236. [Google Scholar] [CrossRef]
- Park, D.; Larson, A.M.; Klibanov, A.M.; Wang, Y. Antiviral and Antibacterial Polyurethanes of Various Modalities. Appl. Biochem. Biotechnol. 2013, 169, 1134–1146. [Google Scholar] [CrossRef] [Green Version]
- Galdiero, S.; Falanga, A.; Vitiello, M.; Cantisani, M.; Marra, V.; Galdiero, M. Silver Nanoparticles as Potential Antiviral Agents. Molecules 2011, 16, 8894–8918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Gusseme, B.; Sintubin, L.; Baert, L.; Thibo, E.; Hennebel, T.; Vermeulen, G.; Uyttendaele, M.; Verstraete, W.; Boon, N. Biogenic Silver for Disinfection of Water Contaminated with Viruses. Appl. Environ. Microbiol. 2009, 76, 1082–1087. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Chen, L.; Duan, S.-M.; Yang, Q.-X.; Yang, M.; Gao, C.; Zhang, B.-Y.; He, H.; Dong, X.-P. Efficient and quick inactivation of SARS coronavirus and other microbes exposed to the surfaces of some metal catalysts. Biomed. Environ. Sci. 2005, 18, 176–180. [Google Scholar] [PubMed]
- Kumar, S.V.; Bafana, A.; Pawar, P.; Faltane, M.; Rahman, A.; Dahoumane, S.; Kucknoor, A.; Jeffryes, C.S. Optimized production of antibacterial copper oxide nanoparticles in a microwave-assisted synthesis reaction using response surface methodology. Colloids Surfaces A Physicochem. Eng. Asp. 2019, 573, 170–178. [Google Scholar] [CrossRef]
- Nanomaterial Surface Coatings Could Prevent COVID-19 Spread. Available online: https://aabgu.org/nano-surface-coating-covid-19/ (accessed on 30 July 2020).
- Weiss, C.; Carriere, M.; Fusco, L.; Capua, I.; Regla-Nava, J.A.; Pasquali, M.; Scott, J.A.; Vitale, F.; Unal, M.A.; Mattevi, C.; et al. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano 2020, 14, 6383–6406. [Google Scholar] [CrossRef]
- StatNano Technology Against COVID-19: Nano Insights into Prevention, Diagnosis, and Treatment. Available online: https://statnano.com/technology-against-covid-19-nano-insights (accessed on 30 July 2020).
- Kim, S.H.; Kwak, S.-Y.; Sohn, B.-H.; Park, T.H. Design of TiO2 nanoparticle self-assembled aromatic polyamide thin-film-composite (TFC) membrane as an approach to solve biofouling problem. J. Membr. Sci. 2003, 211, 157–165. [Google Scholar] [CrossRef]
- Lee, J.E.; Ko, G. Norovirus and MS2 inactivation kinetics of UV-A and UV-B with and without TiO2. Water Res. 2013, 47, 5607–5613. [Google Scholar] [CrossRef]
- Foster, H.; Ditta, I.B.; Varghese, S.; Steele, A. Photocatalytic disinfection using titanium dioxide: Spectrum and mechanism of antimicrobial activity. Appl. Microbiol. Biotechnol. 2011, 90, 1847–1868. [Google Scholar] [CrossRef]
- Hajkova, P.; Spatenka, P.; Horsky, J.; Horska, I.; Kolouch, A. Photocatalytic Effect of TiO2 Films on Viruses and Bacteria. Plasma Process. Polym. 2007, 4, S397–S401. [Google Scholar] [CrossRef]
- Kühn, K.P.; Chaberny, I.; Massholder, K.; Stickler, M.; Benz, V.W.; Sonntag, H.-G.; Erdinger, L. Disinfection of surfaces by photocatalytic oxidation with titanium dioxide and UVA light. Chemosphere 2003, 53, 71–77. [Google Scholar] [CrossRef]
- Sunada, K.; Watanabe, T.; Hashimoto, K. Bactericidal Activity of Copper-Deposited TiO2 Thin Film under Weak UV Light Illumination. Environ. Sci. Technol. 2003, 37, 4785–4789. [Google Scholar] [CrossRef] [PubMed]
- Zan, L.; Fa, W.; Peng, T.; Gong, Z.-K. Photocatalysis effect of nanometer TiO2 and TiO2-coated ceramic plate on Hepatitis B virus. J. Photochem. Photobiol. B Biol. 2007, 86, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Ditta, I.B.; Steele, A.; Liptrot, C.; Tobin, J.; Tyler, H.; Yates, H.M.; Sheel, D.W.; Foster, H. Photocatalytic antimicrobial activity of thin surface films of TiO2, CuO and TiO2/CuO dual layers on Escherichia coli and bacteriophage T4. Appl. Microbiol. Biotechnol. 2008, 79, 127–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashige, N.; Kakita, Y.; Nakashima, Y.; Miake, F.; Watanabe, K. Mechanism of the photocatalytic inactivation of Lactobacillus casei phage PL-1 by titania thin film. Curr. Microbiol. 2001, 42, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Kakita, Y.; Obuchi, E.; Nakano, K.; Murata, K.; Kuroiwa, A.; Miake, F.; Watanabe, K. Photocatalytic Inactivation of Lactobacillus PL-1 Phages by a Thin Film of Titania. Biocontrol Sci. 2000, 5, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.C.; Ho, W.; Lin, J.; Yip, H.; Wong, P.K. Photocatalytic Activity, Antibacterial Effect, and Photoinduced Hydrophilicity of TiO2Films Coated on a Stainless Steel Substrate. Environ. Sci. Technol. 2003, 37, 2296–2301. [Google Scholar] [CrossRef]
- Kwak, S.-Y.; Kim, S.H.; Kim, S.S. Hybrid organic/inorganic reverse osmosis (RO) membrane for bactericidal anti-fouling. Preparation and characterization of TiO2 nanoparticle self-assembled aromatic polyamide thin-film-composite (TFC) membrane. Environ. Sci. Technol. 2001, 35, 2388–2394. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, V.; Papi, M.; Conti, C.; Ciasca, G.; Maulucci, G.; De Spirito, M. The future development of bacteria fighting medical devices: The role of graphene oxide. Expert Rev. Med. Devices 2016, 13, 1013–1019. [Google Scholar] [CrossRef]
- Zamora-Ledezma, C.; Puech, N.; Zakri, C.; Grelet, E.; Moulton, S.E.; Wallace, G.G.; Gambhir, S.; Blanc, C.; Anglaret, E.; Poulin, P. Liquid Crystallinity and Dimensions of Surfactant-Stabilized Sheets of Reduced Graphene Oxide. J. Phys. Chem. Lett. 2012, 3, 2425–2430. [Google Scholar] [CrossRef]
- Wikramaratna, P.; Paton, R.S.; Ghafari, M.; Lourenco, J. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. Epidemiology 2020. [Google Scholar] [CrossRef]
- Younes, N.; Al-Sadeq, D.W.; Al-Jighefee, H.; Younes, S.; Al-Jamal, O.; Daas, H.I.; Yassine, H.M.; Nasrallah, G.K. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2. Viruses 2020, 12, 582. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Luz, P.M.; Wakimoto, M.D.; Veloso, V.G.; Grinsztejn, B.; Perazzo, H. COVID-19: A meta-analysis of diagnostic test accuracy of commercial assays registered in Brazil. Braz. J. Infect. Dis. 2020, 24, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Lee, C.K.; Lam, L.T.M.; Yan, B.; Chua, Y.X.; Lim, A.Y.N.; Phang, K.F.; Kew, G.S.; Teng, H.; Ngai, C.H.; et al. Covert COVID-19 and false-positive dengue serology in Singapore. Lancet Infect. Dis. 2020, 20, 536. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.N.; Kessel, B. False Positives in Reverse Transcription PCR Testing For SARS-CoV-2. Epidemiology 2020. Available online: http://medrxiv.org/lookup/doi/10.1101/2020.04.26.20080911 (accessed on 27 July 2020). [CrossRef]
- Woloshin, S.; Patel, N.; Kesselheim, A.S. False Negative Tests for SARS-CoV-2 Infection—Challenges and Implications. N. Engl. J. Med. 2020, 383, e38. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef] [Green Version]
- Nghiem, L.D.; Morgan, B.; Donner, E.; Short, M.D. The COVID-19 pandemic: Considerations for the waste and wastewater services sector. Case Stud. Chem. Environ. Eng. 2020, 1, 100006. [Google Scholar] [CrossRef]
- Gundy, P.M.; Gerba, C.P.; Pepper, I.L. Survival of Coronaviruses in Water and Wastewater. Food Environ. Virol. 2008, 1, 10–14. [Google Scholar] [CrossRef] [Green Version]
- Bhowmick, G.; Dhar, D.; Nath, D.; Ghangrekar, M.M.; Banerjee, R.; Das, S.; Chatterjee, J. Coronavirus disease 2019 (COVID-19) outbreak: Some serious consequences with urban and rural water cycle. NPJ Clean Water 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Sanvicens, N.; Pastells, C.; Pascual, N.; Marco, M.-P. Nanoparticle-based biosensors for detection of pathogenic bacteria. TrAC Trends Anal. Chem. 2009, 28, 1243–1252. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.A.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Central Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Zhou, Y.; Ye, J.; Al-Maskri, A.A.A.; Kang, Y.; Zeng, S.; Cai, S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020, 10, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Draz, M.S.; Shafiee, H. Applications of gold nanoparticles in virus detection. Theranostics 2018, 8, 1985–2017. [Google Scholar] [CrossRef] [PubMed]
- Dincau, B.M.; Lee, Y.; Kim, J.-H.; Yeo, W.-H. Recent Advances in Nanoparticle Concentration and Their Application in Viral Detection Using Integrated Sensors. Sensors 2017, 17, 2316. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef]
- Galdiero, S.; Rai, M.; Gade, A.; Falanga, A.; Incoronato, N.; Russo, L.; Galdiero, M.; Gaikwad, S.; Ingle, A. Antiviral activity of mycosynthesized silver nanoparticles against herpes simplex virus and human parainfluenza virus type 3. Int. J. Nanomed. 2013, 8, 4303–4314. [Google Scholar] [CrossRef] [Green Version]
- Howes, P.D.; Chandrawati, R.; Stevens, M.M. Colloidal nanoparticles as advanced biological sensors. Science 2014, 346, 1247390. [Google Scholar] [CrossRef] [Green Version]
- Sportelli, M.C.; Izzi, M.; Kukushkina, E.A.; Hossain, S.I.; Picca, R.A.; DiTaranto, N.; Cioffi, N. Can Nanotechnology and Materials Science Help the Fight against SARS-CoV-2? Nanomaterials 2020, 10, 802. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Pei, H.; Yang, F.; Fan, C.; Zuo, X. Applications of Gold Nanoparticles in the Detection and Identification of Infectious Diseases and Biothreats. Adv. Mater. 2013, 25, 3490–3496. [Google Scholar] [CrossRef]
- Mokhtarzadeh, A.; Eivazzadeh-Keihan, R.; Pashazadeh, P.; Hejazi, M.; Gharaatifar, N.; Hasanzadeh, M.; Baradaran, B.; De La Guardia, M. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. 2017, 97, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Tallury, P.; Payton, K.; Santra, S. Silica-based multimodal/multifunctional nanoparticles for bioimaging and biosensing applications. Nanomedicine 2008, 3, 579–592. [Google Scholar] [CrossRef]
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-González, E.; Santiago, G.T.-D.; Lara-Mayorga, I.M.; Martínez-Chapa, S.O.; Alvarez, M.M. Portable and accurate diagnostics for COVID-19: Combined use of the miniPCR thermocycler and a well-plate reader for SARS-CoV-2 virus detection. PLoS ONE 2020, 15, e0237418. [Google Scholar] [CrossRef] [PubMed]
- González-González, E.; Lara-Mayorga, I.M.; García-Rubio, A.; Garciaméndez-Mijares, C.E.; Emilio-Guerra-Alvarez, G.; García, G.; Aguayo, J.; Zhang, Y.S.; Martínez-Chapa, S.O.; Santiago, T.; et al. Scaling diagnostics in times of COVID-19: Rapid prototyping of 3D-printed water circulators for Loop-mediated Isothermal Amplification (LAMP) and detection of SARS-CoV-2 virus. medRxiv 2020, 35. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Du, R.-H.; Li, B.; Zheng, X.-S.; Yang, X.-L.; Hu, B.; Wang, Y.-Y.; Xiao, G.-F.; Yan, B.; Shi, Z.-L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [Green Version]
- Dalum, S.; Hedegård, P. Theory of Chiral Induced Spin Selectivity. Nano Lett. 2019, 19, 5253–5259. [Google Scholar] [CrossRef]
- Naaman, R.; Waldeck, D.H. Spintronics and Chirality: Spin Selectivity in Electron Transport through Chiral Molecules. Annu. Rev. Phys. Chem. 2015, 66, 263–281. [Google Scholar] [CrossRef]
- Aragonès, A.C.; Medina, E.; Gimeno, N.; Teixidó, M.; Palma, J.L.; Tao, N.; Ugalde, J.M.; Huerta, M.F.; Giralt, E.; Díez-Pérez, I.; et al. Measuring the Spin-Polarization Power of a Single Chiral Molecule. Small 2016, 13, 1602519. [Google Scholar] [CrossRef]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020, S1684118220300827. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S. Understanding the Nature of Variations in Structural Sequences Coding for Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N) Proteins of SARS-CoV-2. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3562504 (accessed on 21 August 2020).
- Posthuma-Trumpie, G.A.; Korf, J.; Van Amerongen, A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2008, 393, 569–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, H.V.; Dantzler, J.L.; Weigl, B.H. Analytical Tools to Improve Optimization Procedures for Lateral Flow Assays. Diagnostics 2017, 7, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, W.C.; Beni, V.; Turner, A.P. Lateral-flow technology: From visual to instrumental. TrAC Trends Anal. Chem. 2016, 79, 297–305. [Google Scholar] [CrossRef]
- Oberfeld, B.; Achanta, A.; Carpenter, K.; Chen, P.; Gilette, N.M.; Langat, P.; Said, J.T.; Schiff, A.E.; Zhou, A.S.; Barczak, A.K.; et al. SnapShot: COVID-19. Cell 2020, 181, 954–954.e1. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Lin, C.; Zhang, J.; Oo, M.K.K.; Fan, X. Rapid and quantitative detection of COVID-19 markers in micro-liter sized samples. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.; Lu, Y.; Zhang, J.; Wang, J.; Dan, Y.; Tan, Z.; He, X.; Qian, C.; Sun, Q.; Hu, Q.; et al. Viral Kinetics and Antibody Responses in Patients with COVID-19. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Wang, M.; Zuo, Z.; Fan, C.; Ye, F.; Cai, Z.; Wang, Y.; Cui, H.; Pan, K.; Xu, A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease. Int. J. Infect. Dis. 2020, 94, 49–52. [Google Scholar] [CrossRef]
- Cai, X.-F.; Chen, J.; Hu, J.-L.; Long, Q.-X.; Deng, H.-J.; Liu, P.; Fan, K.; Liao, P.; Liu, B.-Z.; Wu, G.-C.; et al. A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease. J. Infect. Dis. 2020, 222, 189–193. [Google Scholar] [CrossRef]
- Ashtari, P.; He, X.; Zou, Z.; Gong, P. An efficient method for recovery of target ssDNA based on amino-modified silica-coated magnetic nanoparticles. Talanta 2005, 67, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Hung, L.-Y.; Chang, J.-C.; Tsai, Y.-C.; Huang, C.-C.; Chang, C.-P.; Yeh, C.-S.; Lee, G.-B. Magnetic nanoparticle-based immunoassay for rapid detection of influenza infections by using an integrated microfluidic system. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 819–829. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Mou, X.; Deng, Y.; Liu, H.; Li, S.; Xu, L.; Li, X. Conditions of polymerase chain reaction amplification by magnetic enrichment and nanoscale detection sensitivity. J. Nanosci. Nanotechnol. 2012, 12, 3862–3866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, B.; Huang, X.; Bu, P.; Zhuang, J.; Cheng, Z.; Feng, J.; Yang, N.; Dong, C.; Zhang, J.; Yan, X. Influenza virus detection with pentabody-activated nanoparticles. J. Virol. Methods 2010, 169, 282–289. [Google Scholar] [CrossRef]
- Xi, D.; Luo, X.; Lu, Q.; Yao, K.; Liu, Z.; Ning, Q. The detection of HBV DNA with gold-coated iron oxide nanoparticle gene probes. J. Nanopart. Res. 2007, 10, 393–400. [Google Scholar] [CrossRef]
- Torres-Chavolla, E.; Alocilja, E. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef]
- Xiang, J.; Yan, M.; Li, H.; Liu, T.; Lin, C.; Huang, S.; Shen, C. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold- Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Mertens, P.; De Vos, N.; Martiny, D.; Jassoy, C.; Mirazimi, A.; Cuypers, L.; Wijngaert, S.V.D.; Monteil, V.; Melin, P.; Stoffels, K.; et al. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front. Med. 2020, 7, 225. [Google Scholar] [CrossRef]
- Nicoya Application Note: Binding of SARS-CoV-2 Antibody to the Spike Protein Receptor Binding Domain Using OpenSPR-XTTM. Available online: https://nicoyalife.com/blog/sars-cov-2-antibody-binding-kinetics-using-spr/ (accessed on 30 July 2020).
- Bay, J. COVID-19 Impact on Surface Plasmon Resonance Market. Available online: https://www.marketwatch.com/press-release/covid-19-impact-on-surface-plasmon-resonance-market-2020-06-04 (accessed on 30 July 2020).
- Jing, J.-Y.; Wang, Q.; Zhao, W.-M.; Wang, B.-T. Long-range surface plasmon resonance and its sensing applications: A review. Opt. Lasers Eng. 2019, 112, 103–118. [Google Scholar] [CrossRef]
- Mariani, S.; Minunni, M.E. Surface plasmon resonance applications in clinical analysis. Anal. Bioanal. Chem. 2014, 406, 2303–2323. [Google Scholar] [CrossRef]
- PathSensors, Inc. Introducing CANARY—Cutting Edge Pathogen Detection. Available online: https://pathsensors.com/technology/about-canary/ (accessed on 30 July 2020).
- PathSensors, Inc. Announced the Development of a SARS-CoV-2 Biosensor. Available online: https://pathsensors.com/psi-sars-cov-2-biosensor/ (accessed on 30 July 2020).
- Li, T.; Pantazes, R.J.; Maranas, C.D. OptMAVEn—A New Framework for the de novo Design of Antibody Variable Region Models Targeting Specific Antigen Epitopes. PLoS ONE 2014, 9, e105954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapidoth, G.D.; Baran, D.; Pszolla, G.M.; Norn, C.; Alon, A.; Tyka, M.D.; Fleishman, S.J. AbDesign: An algorithm for combinatorial backbone design guided by natural conformations and sequences. Proteins Struct. Funct. Bioinf. 2015, 83, 1385–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schasfoort, R.B.M. Chapter 1. Introduction to Surface Plasmon Resonance. In Handbook of Surface Plasmon Resonance; Schasfoort, R.B.M., Ed.; Royal Society of Chemistry (RSC): Cambridge, UK, 2017; pp. 1–26. [Google Scholar]
- Adolf-Bryfogle, J.; Kalyuzhniy, O.; Kubitz, M.; Weitzner, B.D.; Hu, X.; Adachi, Y.; Schief, W.R.; Dunbrack, R.L. RosettaAntibodyDesign (RAbD): A general framework for computational antibody design. PLoS Comput. Biol. 2018, 14, e1006112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, W.; Kolb, D.M. Density and refractive index of solid layers of noble gases and sulphur hexafluoride. J. Chem. Soc. Faraday Trans. 2 1974, 70, 1098. [Google Scholar] [CrossRef]
- Kreibig, U.; Vollmer, M. Optical Properties of Metal Clusters; Springer Series in Materials Science; Springer: Berlin/Heidelberg, Geramany, 1995; Volume 25, ISBN 978-3-642-08191-0. [Google Scholar]
- Laux, P.; Tentschert, J.; Riebeling, C.; Braeuning, A.; Creutzenberg, O.; Epp, A.; Fessard, V.; Haas, K.-H.; Haase, A.; Hund-Rinke, K.; et al. Nanomaterials: Certain aspects of application, risk assessment and risk communication. Arch. Toxicol. 2017, 92, 121–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yonzon, C.; Stuart, D.; Zhang, X.; McFarland, A.; Haynes, C.L.; Van Duyne, R.P. Towards advanced chemical and biological nanosensors—An overview. Talanta 2005, 67, 438–448. [Google Scholar] [CrossRef]
- Haynes, C.L.; Van Duyne, R.P. Plasmon-Sampled Surface-Enhanced Raman Excitation Spectroscopy. J. Phys. Chem. B 2003, 107, 7426–7433. [Google Scholar] [CrossRef]
- Kim, H.; Park, M.; Hwang, J.; Kim, J.H.; Chung, D.-R.; Lee, K.-S.; Kang, M. Development of Label-Free Colorimetric Assay for MERS-CoV Using Gold Nanoparticles. ACS Sens. 2019, 4, 1306–1312. [Google Scholar] [CrossRef]
- Li, H.; Rothberg, L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 14036–14039. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.C.; Chang, Y.-F.; Chen, K.-H.; Su, L.-C.; Lee, C.-W.; Chen, C.-C.; Chen, Y.M.A.; Chou, C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009, 25, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Schatz, G.C.; Van Duyne, R.P. Electromagnetic Mechanism of Surface-Enhanced Spectroscopy. In Handbook of Vibrational Spectroscopy; Chalmers, J.M., Griffiths, P.R., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2006; pp. 759–774. ISBN 978-0-471-98847-2. [Google Scholar]
- Li, P.; Long, F.; Chen, W.; Chen, J.; Chu, P.K.; Yu, X.-F. Fundamentals and applications of surface-enhanced Raman spectroscopy–based biosensors. Curr. Opin. Biomed. Eng. 2020, 13, 51–59. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Witkowska, E.; Jagielski, T.; Kamińska, A.; Kowalska, A.; Hryncewicz-Gwóźdź, A.; Waluk, J. Detection and identification of human fungal pathogens using surface-enhanced Raman spectroscopy and principal component analysis. Anal. Methods 2016, 8, 8427–8434. [Google Scholar] [CrossRef]
- Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Reverse transcription loop-mediated isothermal amplification combined with nanoparticles-based biosensor for diagnosis of COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Paradiso, A.V.; De Summa, S.; Silvestris, N.; Tommasi, S.; Tufaro, A.; De Palma, G.; LaRocca, A.M.V.; Chironna, M.; D’Addabbo, V.; Raffaele, D.; et al. Rapid serological tests have a role in asymptomatic health workers COVID-19 screening. medRxiv 2020. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.; et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef]
- Shen, W.; Li, M.; Wang, B.; Liu, J.; Li, Z.; Jiang, L.; Song, Y. Hierarchical optical antenna: Gold nanoparticle-modified photonic crystal for highly-sensitive label-free DNA detection. J. Mater. Chem. 2012, 22, 8127. [Google Scholar] [CrossRef]
- Martínez-Paredes, G.; González-García, M.B.; Costa-García, A. Genosensor for SARS Virus Detection Based on Gold Nanostructured Screen-Printed Carbon Electrodes. Electroanalysis 2009, 21, 379–385. [Google Scholar] [CrossRef]
- Choi, Y.; Hwang, J.H.; Lee, S.Y. Recent Trends in Nanomaterials-Based Colorimetric Detection of Pathogenic Bacteria and Viruses. Small Methods 2018, 2, 1700351. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Sato, M.; Sato, Y.; Ando, N.; Takayama, T.; Fujita, M.; Ishihara, M. Synthesis and Application of Silver Nanoparticles (Ag NPs) for the Prevention of Infection in Healthcare Workers. Int. J. Mol. Sci. 2019, 20, 3620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen, C.-W.; De Puig, H.; Tam, J.O.; Gómez-Márquez, J.; Bosch, I.; Hamad-Schifferli, K.; Gehrke, L. Multicolored silver nanoparticles for multiplexed disease diagnostics: Distinguishing dengue, yellow fever, and Ebola viruses. Lab Chip 2015, 15, 1638–1641. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hong, M.; Qiu, B.; Wei, X.; Cai, Z.; Chen, Y.; Chen, G. A highly sensitive chemiluminescent metalloimmunoassay for H1N1 influenza virus detection based on a silver nanoparticle label. Chem. Commun. 2013, 49, 10563–10565. [Google Scholar] [CrossRef]
- Sha, M.Y.; Yamanaka, M.; Walton, I.D.; Norton, S.M.; Stoermer, R.L.; Keating, C.D.; Natan, M.J.; Penn, S.G. Encoded Metal Nanoparticle-Based Molecular Beacons for Multiplexed Detection of DNA. NanoBiotechnology 2005, 1, 327–336. [Google Scholar] [CrossRef]
- Ishikawa, F.N.; Chang, H.-K.; Curreli, M.; Liao, H.-I.; Olson, C.A.; Chen, P.-C.; Zhang, R.; Roberts, R.W.; Sun, R.; Cote, R.J.; et al. Label-Free, Electrical Detection of the SARS Virus N-Protein with Nanowire Biosensors Utilizing Antibody Mimics as Capture Probes. ACS Nano 2009, 3, 1219–1224. [Google Scholar] [CrossRef] [Green Version]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Cu, Y.T.H.; Luo, D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat. Biotechnol. 2005, 23, 885–889. [Google Scholar] [CrossRef]
- Roh, C.; Jo, S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011, 86, 1475–1479. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Q.; Shan, G.; Wang, C.; Du, J.; Wang, S.; Li, Y.; Chen, X.; Jing, X.; Wei, Y. Preparation of silver nanoparticles dispersed in polyacrylonitrile nanofiber film spun by electrospinning. Mater. Lett. 2005, 59, 3046–3049. [Google Scholar] [CrossRef]
- Nicolini, A.M.; McCracken, K.E.; Yoon, J.-Y. Future developments in biosensors for field-ready Zika virus diagnostics. J. Biol. Eng. 2017, 11, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saylan, Y.; Erdem, Ö.; Ünal, S.; Denizli, A. An Alternative Medical Diagnosis Method: Biosensors for Virus Detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pashchenko, O.; Shelby, T.; Banerjee, T.; Santra, S. A Comparison of Optical, Electrochemical, Magnetic, and Colorimetric Point-of-Care Biosensors for Infectious Disease Diagnosis. ACS Infect. Dis. 2018, 4, 1162–1178. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z.; Gittens, M.; Pocock, J.; Tothill, I.E. Biosensors for waterborne viruses: Detection and removal. Biochimie 2015, 115, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.S.; Toh, C.-S. Novel biosensing methodologies for ultrasensitive detection of viruses. Analyst 2013, 138, 6219. [Google Scholar] [CrossRef] [PubMed]
- Pant, S.; Singh, M.; Ravichandiran, V.; Murty, U.S.N.; Srivastava, H.K. Peptide-like and small-molecule inhibitors against Covid-19. J. Biomol. Struct. Dyn. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Choudhir, G.; Shukla, S.K.; Sharma, M.; Tyagi, P.; Bhushan, A.; Rathore, M. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J. Biomol. Struct. Dyn. 2020, 1–11. [Google Scholar] [CrossRef]
- Lim, T.S. (Ed.) Recombinant Antibodies for Infectious Diseases. In Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2017; Volume 1053, ISBN 978-3-319-72076-0. [Google Scholar]
- Coico, R.; Sunshine, G. Immunology: A Short Course, 7th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2015; ISBN 111839691X. [Google Scholar]
- Kuroda, D.; Shirai, H.; Jacobson, M.P.; Nakamura, H. Computer-aided antibody design. Protein Eng. Des. Sel. 2012, 25, 507–522. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, T. Toward rational antibody design: Recent advancements in molecular dynamics simulations. Int. Immunol. 2018, 30, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Kuroda, D.; Kiyoshi, M.; Nakakido, M.; Nagatoishi, S.; Soga, S.; Shirai, H.; Tsumoto, K. Exploring designability of electrostatic complementarity at an antigen-antibody interface directed by mutagenesis, biophysical analysis, and molecular dynamics simulations. Sci. Rep. 2019, 9, 4482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugita, Y.; Okamotoab, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinf. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, J.A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theor. Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; van Buuren, A.R.; Apol, E.; Meulenhoff, P.J.; Tieleman, D.P.; Sijbers, A.L.T.M.; Feenstra, K.A.; van Drunen, R.; et al. Gromacs. User Manual Version 3.2. Available online: http://www.gromacs.org (accessed on 1 January 2013).
- SANDIA LAMMPS. Available online: http://lammps.sandia.gov (accessed on 27 July 2020).
- Whitelegg, N.R.; Rees, A.R. WAM: An improved algorithm for modelling antibodies on the WEB. Protein Eng. Des. Sel. 2000, 13, 819–824. [Google Scholar] [CrossRef] [Green Version]
- Lepore, R.; Olimpieri, P.P.; Messih, M.A.; Tramontano, A. PIGSPro: Prediction of immunoGlobulin structures v2. Nucleic Acids Res. 2017, 45, W17–W23. [Google Scholar] [CrossRef]
- Sircar, A.; Kim, E.T.; Gray, J.J. RosettaAntibody: Antibody variable region homology modeling server. Nucleic Acids Res. 2009, 37, W474–W479. [Google Scholar] [CrossRef] [Green Version]
- Desautels, T.; Zemla, A.K.; Lau, E.Y.; Franco, M.; Faissol, D. Rapid in silico design of antibodies targeting SARS-CoV-2 using machine learning and supercomputing. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Zeng, H.; Mueller, J.; Carter, B.; Wang, Z.; Schilz, J.; Horny, G.; Birnbaum, M.E.; Ewert, S.; Gifford, D.K. Antibody complementarity determining region design using high-capacity machine learning. Bioinformatics 2019, 36, 2126–2133. [Google Scholar] [CrossRef]
- Bragazzi, N.L.; Dai, H.; Damiani, G.; Behzadifar, M.; Martini, M.; Wu, J. How Big Data and Artificial Intelligence Can Help Better Manage the COVID-19 Pandemic. Int. J. Environ. Res. Public Health 2020, 17, 3176. [Google Scholar] [CrossRef]
- Heaven, W.D. Our Weird Behavior during the Pandemic Is Messing with AI Models. Available online: https://www.technologyreview.com/2020/05/11/1001563/covid-pandemic-broken-ai-machine-learning-amazon-retail-fraud-humans-in-the-loop/ (accessed on 3 August 2020).
- Bayram, M.; Springer, S.; Garvey, C.K.; Özdemir, V. COVID-19 Digital Health Innovation Policy: A Portal to Alternative Futures in the Making. OMICS J. Integr. Biol. 2020, 24, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zeng, Z.; Wang, K.; Wong, S.-S.; Liang, W.; Zanin, M.; Liu, P.; Cao, X.; Gao, Z.; Mai, Z.; et al. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J. Thorac. Dis. 2020, 12, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Smits, H.; Knoops, A.J.G.; Korst, M.B.J.M.; Samson, T.; Scholten, E.T.; Schalekamp, S.; Schaefer-Prokop, C.M.; Philipsen, R.H.H.M.; Meijers, A.; et al. COVID-19 on Chest Radiographs: A Multireader Evaluation of an Artificial Intelligence System. Radiology 2020, 296, E166–E172. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, D.; Dong, J.; Wang, N.; Huang, H.; Xu, H.; Xia, C. False-Negative Results of Real-Time Reverse-Transcriptase Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus 2: Role of Deep-Learning-Based CT Diagnosis and Insights from Two Cases. Korean J. Radiol. 2020, 21, 505–508. [Google Scholar] [CrossRef]
- McCall, B. COVID-19 and artificial intelligence: Protecting health-care workers and curbing the spread. Lancet Digit. Health 2020, 2, e166–e167. [Google Scholar] [CrossRef]
- Carlaw, S. Impact on biometrics of Covid-19. Biom. Technol. Today 2020, 2020, 8–9. [Google Scholar] [CrossRef]
- Jean, S.-S.; Lee, P.-I.; Hsueh, P.-R. Treatment options for COVID-19: The reality and challenges. J. Microbiol. Immunol. Infect. 2020, 53, 436–443. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, Z.; Li, J.-W.; Zhao, H.; Wang, G.-Q. Cytokine release syndrome in severe COVID-19: Interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents 2020, 55, 105954. [Google Scholar] [CrossRef]
- Sallard, E.; Lescure, F.-X.; Yazdanpanah, Y.; Mentre, F.; Peiffer-Smadja, N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020, 178, 104791. [Google Scholar] [CrossRef]
- Tse, L.V.; Meganck, R.M.; Graham, R.L.; Baric, R.S. The Current and Future State of Vaccines, Antivirals and Gene Therapies Against Emerging Coronaviruses. Front. Microbiol. 2020, 11, 658. [Google Scholar] [CrossRef]
- Zhao, J.; Alshukairi, A.N.; Baharoon, S.A.; Ahmed, W.A.; Bokhari, A.A.; Nehdi, A.M.; Layqah, L.A.; Alghamdi, M.G.; Al Gethamy, M.M.; Dada, A.M.; et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017, 2, eaan5393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padron-Regalado, E. Vaccines for SARS-CoV-2: Lessons from Other Coronavirus Strains. Infect. Dis. Ther. 2020, 9, 255–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Microbiology 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Liu, M.A. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Draft Landscape of COVID-19 Candidate Vaccines. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 3 August 2020).
- Cohen, J. COVID-19 Vaccine Protects Monkeys From New Coronavirus, Chinese Biotech Reports. Available online: https://www.sciencemag.org/news/2020/04/covid-19-vaccine-protects-monkeys-new-coronavirus-chinese-biotech-reports (accessed on 3 August 2020).
- Zhong, X.; Yang, H.; Guo, Z.-F.; Sin, W.-Y.F.; Chen, W.; Xu, J.; Fu, L.; Wu, J.; Mak, C.-K.G.; Cheng, C.-S.S.; et al. B-Cell Responses in Patients Who Have Recovered from Severe Acute Respiratory Syndrome Target a Dominant Site in the S2 Domain of the Surface Spike Glycoprotein. J. Virol. 2005, 79, 3401–3408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [Green Version]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [Green Version]
- Modjarrad, K.; Roberts, C.C.; Mills, K.T.; Castellano, A.R.; Paolino, K.; Muthumani, K.; Reuschel, E.L.; Robb, M.L.; Racine, T.; Oh, M.-D.; et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: A phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019, 19, 1013–1022. [Google Scholar] [CrossRef] [Green Version]
- Moderna Doses First Patient With mRNA-1273 in Coronavirus Vaccine Trial. Available online: https://www.pharmaceutical-business-review.com/news/moderna-mrna-1273-coronavirus-trial/ (accessed on 3 August 2020).
- Martens, S.L.; Klein, S.; Barnes, R.A.; TrejoSanchez, P.; Roth, C.C.; Ibey, B.L. 600-ns pulsed electric fields affect inactivation and antibiotic susceptibilities of Escherichia coli and Lactobacillus acidophilus. AMB Express 2020, 10, 1–11. [Google Scholar] [CrossRef]













| Innovation | Description | Source | Type |
|---|---|---|---|
| Intensive care unit (ICU) from containers | Shipping containers repurposed to create biocontainment ICU units. | [39] | Medical facility |
| Stanford Pneumask | Reusable full-face snorkel mask | [40] | Personal Protection Equipment (PPE) |
| Maker Mask | Three-dimensional (3D) printable respirator quality masks | [41] | PPE |
| 3D shield | 3D printed powered air purifying respirator (PAPR) created at Duke University | [42] | PAPR |
| Massachusetts Institute of Technology (MIT) origami face shield | Single piece disposable face shields for mass production | [43] | PPE |
| Epidax | National University of Singapore portable on-site COVID-19 polymerase chain reaction (PCR) diagnostic system | [44] | Diagnostic test |
| Glassafe | Transparent material to make close proximity safer. Used in aircraft to isolate passenger seats. | [45] | Social distancing |
| MIT Emergency ventilator | Open-source, low-cost ventilator | [46] | Ventilator |
| Resuscitation bags low-cost ventilator | Resuscitation bag adaptation to build an emergency low cost ventilator (Georgia Tech) | [47] | Ventilator |
| Ventilator Intervention Technology Accessible Locally (US National Aeronautics and Space Administration (NASA) VITAL Ventilator) | Emergency ventilator prototype developed in 37 days | [48] | Ventilator |
| Harvard-MIT detection mask | Face mask that lights up a fluorescent signal in the presence of SARS-CoV-2 | [49] | PPE and diagnostic |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zamora-Ledezma, C.; C., D.F.C.; Medina, E.; Sinche, F.; Santiago Vispo, N.; Dahoumane, S.A.; Alexis, F. Biomedical Science to Tackle the COVID-19 Pandemic: Current Status and Future Perspectives. Molecules 2020, 25, 4620. https://doi.org/10.3390/molecules25204620
Zamora-Ledezma C, C. DFC, Medina E, Sinche F, Santiago Vispo N, Dahoumane SA, Alexis F. Biomedical Science to Tackle the COVID-19 Pandemic: Current Status and Future Perspectives. Molecules. 2020; 25(20):4620. https://doi.org/10.3390/molecules25204620
Chicago/Turabian StyleZamora-Ledezma, Camilo, David F. Clavijo C., Ernesto Medina, Federico Sinche, Nelson Santiago Vispo, Si Amar Dahoumane, and Frank Alexis. 2020. "Biomedical Science to Tackle the COVID-19 Pandemic: Current Status and Future Perspectives" Molecules 25, no. 20: 4620. https://doi.org/10.3390/molecules25204620
APA StyleZamora-Ledezma, C., C., D. F. C., Medina, E., Sinche, F., Santiago Vispo, N., Dahoumane, S. A., & Alexis, F. (2020). Biomedical Science to Tackle the COVID-19 Pandemic: Current Status and Future Perspectives. Molecules, 25(20), 4620. https://doi.org/10.3390/molecules25204620