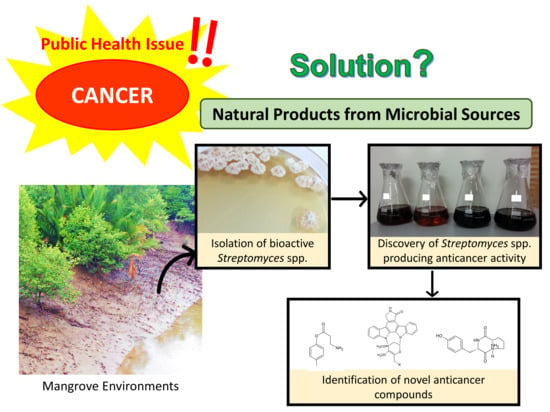
Anticancer Drug Discovery from Microbial Sources: The Unique Mangrove Streptomycetes
Abstract
:1. Introduction
2. Microbial Sources: Why the Streptomyces?
3. Bioprospecting of Streptomyces spp. from Mangrove Environment
4. Discovery of Novel Compounds with Anticancer/Cytotoxic Activity from Mangrove-Derived Streptomyces spp.
5. Crude Extracts of Mangrove-Derived Streptomyces spp. with Anticancer/Cytotoxic Activity
| No. | Author | Strain | Source | Compound/Crude Extract | IC50 (µg/mL) |
|---|---|---|---|---|---|
| Pure Compound | |||||
| 1. | Xie et al. [99] | Streptomyces sp. 124092 | Mangrove rhizosphere soil of Heritiera littoralis (China) |
| SMMC-7721 cells: (1) 22.0; (2) 60.0; (3) 13.6; (4) 6.0 |
| 2. | Yuan et al. [100] | Streptomyces sp. 211726 | Mangrove rhizosphere soil of Heritiera globosa (China) |
| HCT-116 cells: (1) 5.64; (2) 2.58 |
| 3. | Fu et al. [101] | Streptomyces sp. FMA | Mangrove soil (China) |
| HL-60 cells: (1) 0.67 a A549 cells: (1) 2.41 a HeLa cells: (1) 16.61 a; (2) 10.47 a |
| 4. | Cibi and Nair [132] | Streptomyces parvulus CBJ1 | Mangrove soil (India) | Actinomycin D | A549 cells: 0.52 |
| 5. | Fu et al. [124] | Streptomyces antibioticus strain H74-21 | Sediment from a mangrove site (China) | Streptomyceamide C * | MCF-7 cells: 27.0 |
| 6. | Mangamuri et al. [125] | Streptomyces cheonanensis VUK-A | Sediment from Coringa mangrove ecosystem (India) | 2-Methyl butyl propyl phthalate * | MCF-7 cells: 1391.7 MDA-MB-231 cells: 278.34 HeLa cells: 27.834 OAW-42 cells: 278.34 |
| 7. | Zhang et al. [123] | Streptomyces anandii H41-59 | Sediment from a mangrove district (China) |
5β,6β-epoxy-ergosta-8(14),22-diene-3β,7β-diol | MCF-7 cells: all compounds exhibited > 50; except (3) 18.1, (8) 24.3, (10) 17.3, and (11) 27.4 SF-268 cells: all compounds exhibited > 50; except (3) 13.0, (8) 15.5, (10) 27.8, and (11) 25.1 NCl-H460 cells: all compounds exhibited > 50; except (3) 23.5, (8) 19.8, (10) 23.7, and (11) 23.7 |
| 8. | Han et al. [102] | Streptomyces sp. 219807 | Mangrove soil (China) |
| HeLa cells: (1) 0.27 a; (2) 1.20 a; (3) 0.30 a; (4) 0.30 a; (5) 0.59 a; (6) 0.60 a; (7) 0.60 a MCF-7 cells: (1) 0.30 a; (2) 2.26 a; (3) 0.30 a; (4) 0.19 a; (5) 1.00 a; (6) 1.01 a; (7) 0.80 a |
| 9. | Hu et al. [119] | Streptomyces antibioticus H12-15 | Sediment from a mangrove district (China) |
| MCF-7 cells: (1) >50; (2) >50; (3) 18.1; (4) 36.4; (5) 26.1 SF-268 cells: (1) 33.6; (2) 41.6; (3) <1.6; (4) <1.6; (5) <1.6 NCl-H460 cells: (1) >50; (2) >50; (3) 21.7; (4) 43.7; (5) 15.5 |
| Crude Extract | |||||
| 10. | Ravikumar et al. [126] | Streptomyces sp. ACT01, ACT02, ACT03, ACT04, and ACT 05 | Mangrove sediment from Manakkudi mangrove ecosystem (India) | Crude ethyl acetate extract | MCF-7 cells: (ACT01) 19.49; (ACT02) 29.94; (ACT03) 75.54; (ACT04) 66.04; (ACT05) 92.64 MDA-MB-231 cells: (ACT01) 32.79; (ACT02) 69.84; (ACT03) 84.09; (ACT04) >100; (ACT05) >100 |
| 11. | Ser et al. [33] | Streptomyces pluripotens MUSC 137T | Mangrove soil (Malaysia) | Crude methanol extract | MCF-7 cells: 61.33 HCT-116 cells: 83.72 A549 cells: 147.20 Ca Ski cells: 300.50 HT-29 cells: 300.98 Caco-2, SW480 and DU145 cells: >400 |
| 12. | Tan et al. [29] | Streptomyces MUM256 | Mangrove soil (Malaysia) | Crude methanol extract | HCT-116 cells: 292.33 HT-29, Caco-2, SW480, MCF-7, A549, DU 145, and Ca Ski cells: b N.P |
| 13. | Sanjivkumar et al. [133] | Streptomyces olivaceus (MSU3) | Mangrove rhizosphere soil of Rhizophora mucronata (India) | Crude ethyl acetate extract | MCF-7 cells: 88.26 HT-29 cells: 104.81 |
| 14. | Ser et al. [30] | Streptomyces malaysiense MUSC 136T | Mangrove soil (Malaysia) | Crude methanol extract | HCT-116, HT-29, A549, and Ca Ski cells: b N.P. |
| 15. | Law et al. [84] | Streptomyces colonosanans MUSC 93JT | Mangrove soil (Malaysia) | Crude methanol extract | HCT-116, HT-29, Caco-2, and SW480 cells: b N.P. |
| 16. | Ser et al. [134] | Streptomyces gilvigriseus MUSC 26T | Mangrove soil (Malaysia) | Crude methanol extract | HCT-116, HT-29, Caco-2, and SW480 cells: b N.P. |
| 17. | Law et al. [135] | Streptomyces monashensis MUSC 1JT | Mangrove soil (Malaysia) | Crude methanol extract | HCT-116 and SW480 cells: b N.P. |
| 17 | Law et al. [75] | 11 strains of Streptomyces sp. | Mangrove soil (Malaysia) | Crude methanol extract | HCT-116, HT-29, Caco-2, and SW480 cells: b N.P. |
| 18 | Tan et al. [127] | Streptomyces sp. MUM256 | Mangrove soil (Malaysia) | Ethyl acetate extract (further study from Tan et al. [29]) | HCT-116: 88.44 |
| 19 | Tan et al. [3] | Streptomyces sp. MUM 265 | Mangrove soil (Malaysia) | Crude methanol extract | HT29 and Caco-2: b N.P. |
6. Limitations and Suggestions for Future Research
7. Conclusions
Funding
Conflicts of Interest
References
- Are, C.; McMasters, K.M.; Giuliano, A.; Yanala, U.; Balch, C.; Anderson, B.O.; Berman, R.; Audisio, R.; Kovacs, T.; Savant, D. Global forum of cancer surgeons: Perspectives on barriers to surgical care for cancer patients and potential solutions. Ann. Surg. Oncol 2019, 26, 1577–1582. [Google Scholar] [CrossRef]
- Chalbatani, G.M.; Dana, H.; Memari, F.; Gharagozlou, E.; Ashjaei, S.; Kheirandish, P.; Marmari, V.; Mahmoudzadeh, H.; Mozayani, F.; Maleki, A.R. Biological function and molecular mechanism of pirna in cancer. Pract. Lab. Med. 2019, 13, e00113. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T.-H.; Chan, K.-G.; Pusparajah, P.; Yin, W.-F.; Khan, T.M.; Lee, L.-H.; Goh, B.-H. Mangrove derived streptomyces sp. Mum265 as a potential source of antioxidant and anticolon-cancer agents. BMC Microbiol. 2019, 19, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, O.; Zbuk, K. Colorectal cancer in adolescents and young adults: Defining a growing threat. Pediatr. Blood Cancer 2019, 66, e27941. [Google Scholar] [CrossRef] [PubMed]
- Ambrosone, C.B.; Freudenheim, J.L.; Graham, S.; Marshall, J.R.; Vena, J.E.; Brasure, J.R.; Michalek, A.M.; Laughlin, R.; Nemoto, T.; Gillenwater, K.A. Cigarette smoking, n-acetyltransferase 2 genetic polymorphisms, and breast cancer risk. JAMA 1996, 276, 1494–1501. [Google Scholar] [CrossRef] [PubMed]
- Limsui, D.; Vierkant, R.A.; Tillmans, L.S.; Wang, A.H.; Weisenberger, D.J.; Laird, P.W.; Lynch, C.F.; Anderson, K.E.; French, A.J.; Haile, R.W. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J. Natl. Cancer Inst. 2010, 102, 1012–1022. [Google Scholar] [CrossRef]
- Collaboration, G.B.o.D.C. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: A systematic analysis for the global burden of disease study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Pham-Huy, L.A.; He, H.; Pham-Huy, C. Free radicals, antioxidants in disease and health. Int. J. Biomed. Sci. 2008, 4, 89–96. [Google Scholar]
- Clark, R.; Lee, S.-H. Anticancer properties of capsaicin against human cancer. Anticancer Res. 2016, 36, 837–843. [Google Scholar]
- Thoreson, M.A.; Reynolds, A.B. Altered expression of the catenin p120 in human cancer: Implications for tumor progression. Differentiation 2002, 70, 583–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wainwright, E.N.; Scaffidi, P. Epigenetics and cancer stem cells: Unleashing, hijacking, and restricting cellular plasticity. Trends Cancer 2017, 3, 372–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverstein, A.; Silverstein, V.; Nunn, L. Cancer: Conquering a Deadly Disease: Twenty; Twenty-First Century Books: Minneapolis, MN, USA, 2006. [Google Scholar]
- Clark, A.G.; Vignjevic, D.M. Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 2015, 36, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Cui, Q.; Ma, Y.; Jaramillo, M.; Bari, H.; Awan, A.; Yang, S.; Zhang, S.; Liu, L.; Lu, M.; O’Connor-McCourt, M. A map of human cancer signaling. Mol. Syst. Biol. 2007, 3, 152. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Kapoor, K.; Kaur, H. Plants as a source of anticancer agents. J. Nat. Prod. Plant Resour. 2011, 1, 119–124. [Google Scholar]
- Huber, P.E.; Bischof, M.; Jenne, J.; Heiland, S.; Peschke, P.; Saffrich, R.; Gröne, H.-J.; Debus, J.; Lipson, K.E.; Abdollahi, A. Trimodal cancer treatment: Beneficial effects of combined antiangiogenesis, radiation, and chemotherapy. Cancer Res. 2005, 65, 3643–3655. [Google Scholar] [CrossRef] [Green Version]
- Katz, S.J.; Lantz, P.M.; Janz, N.K.; Fagerlin, A.; Schwartz, K.; Liu, L.; Deapen, D.; Salem, B.; Lakhani, I.; Morrow, M. Patient involvement in surgery treatment decisions for breast cancer. J. Clin. Oncol. 2005, 23, 5526–5533. [Google Scholar] [CrossRef]
- Temple, L.K.; Hsieh, L.; Wong, W.D.; Saltz, L.; Schrag, D. Use of surgery among elderly patients with stage iv colorectal cancer. J. Clin. Oncol. 2004, 22, 3475–3484. [Google Scholar] [CrossRef]
- Cragg, G.; Newman, D. Nature: A vital source of leads for anticancer drug development. Phytochem. Rev. 2009, 8, 313–331. [Google Scholar] [CrossRef]
- Da Rocha, A.B.; Lopes, R.M.; Schwartsmann, G. Natural products in anticancer therapy. Curr. Opin. Pharm. 2001, 1, 364–369. [Google Scholar] [CrossRef]
- Simmons, T.L.; Andrianasolo, E.; McPhail, K.; Flatt, P.; Gerwick, W.H. Marine natural products as anticancer drugs. Mol. Cancer Ther. 2005, 4, 333–342. [Google Scholar] [PubMed]
- Gordaliza, M. Natural products as leads to anticancer drugs. Clin. Transl. Oncol. 2007, 9, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Nobili, S.; Lippi, D.; Witort, E.; Donnini, M.; Bausi, L.; Mini, E.; Capaccioli, S. Natural compounds for cancer treatment and prevention. Pharmacol. Res. 2009, 59, 365–378. [Google Scholar] [CrossRef]
- Kinghorn, A.D.; Chin, Y.-W.; Swanson, S.M. Discovery of natural product anticancer agents from biodiverse organisms. Curr. Opin. Drug Discuss. 2009, 12, 189–196. [Google Scholar]
- Anulika, N.P.; Ignatius, E.O.; Raymond, E.S.; Osasere, O.-I.; Abiola, A.H. The chemistry of natural product: Plant secondary metabolites. Int. J. Technol. Enhanc. Emerg. Eng. Res. 2016, 4, 1–8. [Google Scholar]
- Zhang, L.; An, R.; Wang, J.; Sun, N.; Zhang, S.; Hu, J.; Kuai, J. Exploring novel bioactive compounds from marine microbes. Curr. Opin. Microbiol. 2005, 8, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Aravindaram, K.; Yang, N.-S. Anti-inflammatory plant natural products for cancer therapy. Planta Med. 2010, 76, 1103–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.T.-H.; Ser, H.-L.; Yin, W.-F.; Chan, K.-G.; Lee, L.-H.; Goh, B.-H. Investigation of antioxidative and anticancer potentials of streptomyces sp. Mum256 isolated from malaysia mangrove soil. Front. Microbiol. 2015, 6, 1316. [Google Scholar] [CrossRef]
- Ser, H.-L.; Palanisamy, U.D.; Yin, W.-F.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Streptomyces malaysiense sp. Nov.: A novel malaysian mangrove soil actinobacterium with antioxidative activity and cytotoxic potential against human cancer cell lines. Sci. Rep. 2016, 6, 24247. [Google Scholar] [CrossRef] [Green Version]
- Kroschinsky, F.; Stölzel, F.; von Bonin, S.; Beutel, G.; Kochanek, M.; Kiehl, M.; Schellongowski, P. New drugs, new toxicities: Severe side effects of modern targeted and immunotherapy of cancer and their management. Crit. Care 2017, 21, 89. [Google Scholar] [CrossRef] [Green Version]
- Suter, T.M.; Ewer, M.S. Cancer drugs and the heart: Importance and management. Eur. Heart J. 2013, 34, 1102–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ser, H.-L.; Ab Mutalib, N.-S.; Yin, W.-F.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Evaluation of antioxidative and cytotoxic activities of Streptomyces pluripotens musc 137 isolated from mangrove soil in malaysia. Front. Microbiol. 2015, 6, 1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, G.; Gyllenhaal, C.; Soejarto, D. Biodiversity as a source of anticancer drugs. Curr. Drug Targets 2006, 7, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.-J. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer 2003, 3, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, X.; Saravanan, R.; Li, C.M.; Leong, S.S.J. Antimicrobial macromolecules: Synthesis methods and future applications. RSC Adv. 2012, 2, 4031–4044. [Google Scholar] [CrossRef]
- Ab Mutalib, N.-S.; Wong, S.H.; Ser, H.-L.; Duangjai, A.; Law, J.W.-F.; Ratnakomala, S.; Tan, L.T.-H.; Letchumanan, V. Bioprospecting of microbes for valuable compounds to mankind. Prog. Micobes Mol. Biol. 2020, 3, a0000088. [Google Scholar] [CrossRef]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Rayan, A.; Raiyn, J.; Falah, M. Nature is the best source of anticancer drugs: Indexing natural products for their anticancer bioactivity. PLoS ONE 2017, 12, e0187925. [Google Scholar] [CrossRef]
- Martínez-Montiel, N.; Rosas-Murrieta, N.H.; Martínez-Montiel, M.; Gaspariano-Cholula, M.P.; Martínez-Contreras, R.D. Microbial and natural metabolites that inhibit splicing: A powerful alternative for cancer treatment. Biomed Res. Int. 2016, 2016, 3681094. [Google Scholar] [CrossRef] [Green Version]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [Green Version]
- Castelle, C.J.; Banfield, J.F. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 2018, 172, 1181–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, J.W.-F.; Pusparajah, P.; Ab Mutalib, N.-S.; Wong, S.H.; Goh, B.-H.; Lee, L.-H. A review on mangrove actinobacterial diversity: The roles of Streptomyces and novel species discovery. Prog. Micobes Mol. Biol. 2019, 1, a0000024. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.-F.; Letchumanan, V.; Tan, L.T.-H.; Ser, H.-L.; Goh, B.-H.; Lee, L.-H. The rising of “modern actinobacteria” era. Prog. Micobes Mol. Biol. 2020, 3, a0000064. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salam, N.; Jiao, J.-Y.; Zhang, X.-T.; Li, W.-J. Update on the classification of higher ranks in the phylum actinobacteria. Int. J. Syst. Evol. Microbiol. 2020, 70, 1331–1355. [Google Scholar] [CrossRef] [PubMed]
- Dharmaraj, S. Marine streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Lee, L.-H.; Zainal, N.; Azman, A.-S.; Eng, S.-K.; Goh, B.-H.; Yin, W.-F.; Ab Mutalib, N.-S.; Chan, K.-G. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in malaysia. Sci. World J. 2014, 2014, 698178. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.-H.; Zainal, N.; Azman, A.-S.; Eng, S.-K.; Ab Mutalib, N.-S.; Yin, W.-F.; Chan, K.-G. Streptomyces pluripotens sp. Nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 2014, 64, 3297–3306. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.T.-H.; Chan, K.-G.; Khan, T.M.; Bukhari, S.I.; Saokaew, S.; Duangjai, A.; Pusparajah, P.; Lee, L.-H.; Goh, B.-H. Streptomyces sp. Mum212 as a source of antioxidants with radical scavenging and metal chelating properties. Front. Pharmacol. 2017, 8, 276. [Google Scholar] [CrossRef]
- Kemung, H.M.; Hern, T.; Loh, T.; Khan, T.M.; FASc, C.; Gan, K.; Pusparajah, P.; Goh, B.H.; Lee, L.-H. Streptomyces as a prominent resource of future anti-mrsa drugs. Front. Microbiol. 2018, 9, 2221. [Google Scholar] [CrossRef]
- Pimentel-Elardo, S.M.; Kozytska, S.; Bugni, T.S.; Ireland, C.M.; Moll, H.; Hentschel, U. Anti-parasitic compounds from Streptomyces sp. Strains isolated from mediterranean sponges. Mar. Drugs 2010, 8, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.-L.; Law, J.W.-F.; Chaiyakunapruk, N.; Jacob, S.A.; Palanisamy, U.D.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Fermentation conditions that affect clavulanic acid production in Streptomyces clavuligerus: A systematic review. Front. Microbiol. 2016, 7, 522. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, R.; Rong, X.; Huang, Y.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M. Streptomyces bullii sp. Nov., isolated from a hyper-arid atacama desert soil. Antonie Leeuwenhoek 2013, 103, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.-H.; Cheah, Y.-K.; Sidik, S.M.; Ab Mutalib, N.-S.; Tang, Y.-L.; Lin, H.-P.; Hong, K. Molecular characterization of antarctic actinobacteria and screening for antimicrobial metabolite production. World J. Microbiol. Biotechnol. 2012, 28, 2125–2137. [Google Scholar] [CrossRef]
- Tatar, D.; Guven, K.; Spröer, C.; Klenk, H.-P.; Sahin, N. Streptomyces iconiensis sp. Nov. And streptomyces Smyrnaeus sp. Nov., two halotolerant actinomycetes isolated from a salt lake and saltern. Int. J. Syst. Evol. Microbiol. 2014, 64, 3126–3133. [Google Scholar] [CrossRef] [Green Version]
- Flärdh, K.; Buttner, M.J. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009, 7, 36–49. [Google Scholar] [CrossRef]
- Ser, H.-L.; Tan, W.-S.; Ab Mutalib, N.-S.; Cheng, H.-J.; Yin, W.-F.; Chan, K.-G.; Lee, L.-H. Genome sequence of Streptomyces pluripotens musc 135t exhibiting antibacterial and antioxidant activity. Mar. Genom. 2015, 24, 281–283. [Google Scholar] [CrossRef]
- Ser, H.-L.; Chan, K.-G.; Tan, W.-S.; Yin, W.-F.; Goh, B.-H.; Ab Mutalib, N.-S.; Lee, L.-H. Complete genome of mangrove-derived anti-mrsa streptomycete, Streptomyces pluripotens musc 135t. Prog. Micobes Mol. Biol. 2018, 1, a0000004. [Google Scholar] [CrossRef] [Green Version]
- Ser, H.-L.; Tan, W.-S.; Yin, W.-F.; Chan, K.-G.; Ab Mutalib, N.S.; Lee, L.-H. Whole genome sequence of Streptomyces humi strain musc 119t isolated from intertidal soil. Prog. Drug. Discov. Biomed. Sci. 2019, 2, a0000020. [Google Scholar] [CrossRef]
- Ser, H.-L.; Law, J.W.-F.; Tan, W.-S.; Yin, W.-F.; Chan, K.-G. Whole genome sequence of Streptomyces colonosanans strain musc 93jt isolated from mangrove forest in malaysia. Prog. Micobes Mol. Biol. 2020, 3, a0000061. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kasahara, K.; Hirose, Y.; Murakami, K.; Kugimiya, R.; Ochi, K. Activation and products of the cryptic secondary metabolite-biosynthetic gene clusters by rifampicin resistance (rpob) mutations in actinomycetes. J. Bacteriol. 2013, 195, 2959–2970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D. Complete genome sequence of the model actinomycete Streptomyces coelicolor a3 (2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Sakaki, Y.; Hattori, M.; Ōmura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Law, J.W.-F.; Ser, H.-L.; Khan, T.M.; Chuah, L.-H.; Pusparajah, P.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. The potential of streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, L.-H.; Chan, K.-G.; Stach, J.; Wellington, E.M.H.; Goh, B.-H. Editorial: The search for biological active agent(s) from Actinobacteria. Front. Microbiol. 2018, 9, 824. [Google Scholar] [CrossRef] [Green Version]
- Ser, H.-L.; Tan, W.-S.; Mutalib, N.-S.A.; Yin, W.-F.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Genome sequence of Streptomyces gilvigriseus musc 26t isolated from mangrove forest. Braz. J. Microbiol. 2018, 49, 207–209. [Google Scholar] [CrossRef]
- Demain, A.L.; Vaishnav, P. Natural products for cancer chemotherapy. Microb. Biotechnol. 2011, 4, 687–699. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, B.; Mukherjee, S. Cancer therapy using antibiotics. J. Cancer Ther. 2015, 6, 849–858. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.; Morris-Natschke, S.L.; Lee, K.H. Strategies for the optimization of natural leads to anticancer drugs or drug candidates. Med. Res. Rev. 2016, 36, 32–91. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, M.; Fiedler, H.-P. A guide to successful bioprospecting: Informed by actinobacterial systematics. Antonie van Leeuwenhoek 2010, 98, 119–142. [Google Scholar] [CrossRef]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of culturable actinomycetes in hyper-arid soils of the atacama desert, chile. Antonie Leeuwenhoek 2009, 95, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, M.; Pessi, I.S.; Arguelles-Arias, A.; Noirfalise, P.; Luis, G.; Ongena, M.; Barton, H.; Carnol, M.; Rigali, S. Streptomyces lunaelactis sp. Nov., a novel ferroverdin a-producing streptomyces species isolated from a moonmilk speleothem. Antonie van Leeuwenhoek 2015, 107, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.-L.; Ab Mutalib, N.-S.; Yin, W.-F.; Goh, B.-H.; Lee, L.-H.; Chan, K.-G. Genome sequence of streptomyces antioxidans musc 164t isolated from mangrove forest. Prog. Micobes Mol. Biol. 2018, 1, a0000001. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.-F.; Chan, K.-G.; He, Y.-W.; Khan, T.M.; Ab Mutalib, N.-S.; Goh, B.-H.; Lee, L.-H. Diversity of Streptomyces spp. From mangrove forest of sarawak (malaysia) and screening of their antioxidant and cytotoxic activities. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.-F.; Tan, K.-X.; Wong, S.H.; Ab Mutalib, N.-S.; Lee, L.-H. Taxonomic and characterization methods of Streptomyces: A review. Prog. Micobes Mol. Biol. 2018, 1, a0000009. [Google Scholar] [CrossRef] [Green Version]
- Ser, H.-L.; Tan, L.T.-H.; Law, J.W.-F.; Chan, K.-G.; Duangjai, A.; Saokaew, S.; Pusparajah, P.; Ab Mutalib, N.-S.; Khan, T.M.; Goh, B.-H. Focused review: Cytotoxic and antioxidant potentials of mangrove-derived Streptomyces. Front. Microbiol. 2017, 8, 2065. [Google Scholar] [CrossRef]
- Valliappan, K.; Sun, W.; Li, Z. Marine actinobacteria associated with marine organisms and their potentials in producing pharmaceutical natural products. Appl. Microbiol. Biotechnol. 2014, 98, 7365–7377. [Google Scholar] [CrossRef]
- Ser, H.-L.; Palanisamy, U.D.; Yin, W.-F.; Abd Malek, S.N.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Presence of antioxidative agent, pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro-in newly isolated Streptomyces mangrovisoli sp. Nov. Front. Microbiol. 2015, 6, 854. [Google Scholar] [CrossRef] [Green Version]
- Ser, H.-L.; Zainal, N.; Palanisamy, U.D.; Goh, B.-H.; Yin, W.-F.; Chan, K.-G.; Lee, L.-H. Streptomyces Gilvigriseus sp. Nov., a novel actinobacterium isolated from mangrove forest soil. Antonie Leeuwenhoek 2015, 107, 1369–1378. [Google Scholar] [CrossRef]
- Rönnbäck, P.; Crona, B.; Ingwall, L. The return of ecosystem goods and services in replanted mangrove forests: Perspectives from local communities in kenya. Environ. Conserv. 2007, 34, 313–324. [Google Scholar] [CrossRef]
- Ashton, E.C.; Macintosh, D.J. Preliminary assessment of the plant diversity and community ecology of the sematan mangrove forest, sarawak, malaysia. For. Ecol. Manag. 2002, 166, 111–129. [Google Scholar] [CrossRef]
- Sahoo, K.; Dhal, N. Potential microbial diversity in mangrove ecosystems: A review. Indian J. Mar. Sci. 2009, 32, 249–256. [Google Scholar]
- Law, J.W.-F.; Ser, H.-L.; Duangjai, A.; Saokaew, S.; Bukhari, S.I.; Khan, T.M.; Ab Mutalib, N.-S.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Streptomyces colonosanans sp. Nov., a novel actinobacterium isolated from malaysia mangrove soil exhibiting antioxidative activity and cytotoxic potential against human colon cancer cell lines. Front. Microbiol. 2017, 8, 877. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.-L.; Tan, L.T.-H.; Palanisamy, U.D.; Abd Malek, S.N.; Yin, W.-F.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Streptomyces antioxidans sp. Nov., a novel mangrove soil actinobacterium with antioxidative and neuroprotective potentials. Front. Microbiol. 2016, 7, 899. [Google Scholar] [CrossRef] [Green Version]
- Anderson, A.S.; Wellington, E. The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 2001, 51, 797–814. [Google Scholar] [CrossRef] [Green Version]
- Hong, K.; Gao, A.-H.; Xie, Q.-Y.; Gao, H.G.; Zhuang, L.; Lin, H.-P.; Yu, H.-P.; Li, J.; Yao, X.-S.; Goodfellow, M.; et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in china. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef]
- Xu, D.-B.; Ye, W.-W.; Han, Y.; Deng, Z.-X.; Hong, K. Natural products from mangrove actinomycetes. Mar. Drugs 2014, 12, 2590–2613. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Ye, Y.; Wang, R.; Zhang, Y.; Wu, C.; Debnath, S.C.; Ma, Z.; Wang, J.; Wu, M. Streptomyces nigra sp. Nov. Is a novel actinobacterium isolated from mangrove soil and exerts a potent antitumor activity in vitro. Front. Microbiol. 2018, 9, 1587. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Wang, Y.; Luo, Y.; Xie, S.-J.; Ruan, J.-S.; Xu, J. Streptomyces avicenniae sp. Nov., a novel actinomycete isolated from the rhizosphere of the mangrove plant Avicennia mariana. Int. J. Syst. Evolut. Microbiol. 2009, 59, 2624–2628. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, M.; Mitra, A.; Pramanik, A.; Saha, M.; Gachhui, R.; Mukherjee, J. Streptomyces sundarbansensis sp. Nov., an actinomycete that produces 2-allyloxyphenol. Int. J. Syst. Evol. Microbiol. 2011, 61, 2664–2669. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Y.; Xie, S.-J.; Xu, J.; Xiao, J.; Ruan, J.-S. Streptomyces xiamenensis sp. Nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 2009, 59, 472–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Münch, J.; Goerls, H.; Maier, A.; Fiebig, H.-H.; Lin, W.-H.; Hertweck, C. Xiamycin, a pentacyclic indolosesquiterpene with selective anti-hiv activity from a bacterial mangrove endophyte. Bioorg. Med. Chem. Lett. 2010, 20, 6685–6687. [Google Scholar] [CrossRef] [PubMed]
- Kemung, H.M.; Tan, L.T.-H.; Chan, K.-G.; Ser, H.-L.; Law, J.W.-F.; Lee, L.-H.; Goh, B.-H. Investigating the antioxidant potential of Streptomyces sp. Musc 11 from mangrove soil in malaysia. Prog. Drug. Discov. Biomed. Sci. 2019, 2, a0000033. [Google Scholar] [CrossRef]
- Kemung, H.M.; Tan, L.T.-H.; Chan, K.-G.; Ser, H.-L.; Law, J.W.-F.; Lee, L.-H.; Goh, B.-H. Antioxidant activities of streptomyces sp. Strain musc 14 from mangrove forest soil in malaysia. Biomed Res. Int. 2020, 2020, 6402607. [Google Scholar] [CrossRef]
- Kemung, H.M.; Tan, L.T.-H.; Chan, K.-G.; Ser, H.-L.; Law, J.W.-F.; Goh, B.H. Streptomyces sp. Strain musc 5 from mangrove forest in malaysia: Identification, antioxidant potential and chemical profiling of its methanolic extract. Prog. Micobes Mol. Biol. 2020, 3, a0000087. [Google Scholar] [CrossRef]
- Kemung Mangzira, H.; Tan, L.T.-H.; Chan, K.-G.; Ser, H.-L.; Law, J.W.-F.; Lee, L.-H.; Goh, B.-H. Streptomyces sp. Strain musc 125 from mangrove soil in malaysia with anti-mrsa, anti-biofilm and antioxidant activities. Molecules 2020, 25, 3545. [Google Scholar] [CrossRef]
- van Meerloo, J.; Kaspers, G.J.; Cloos, J. Cell sensitivity assays: The mtt assay. In Cancer Cell Culture: Methods and Protocols; Cree, I.A., Ed.; Springer: New York, NY, USA, 2011; pp. 237–245. [Google Scholar]
- Xie, X.-C.; Mei, W.-L.; Zeng, Y.-B.; Lin, H.-P.; Zhuang, L.; Dai, H.-F.; Hong, K. Cytotoxic constituents from marine actinomycete Streptomyces sp. 124092. Chem. J. Chin. Univ. 2008, 29, 2183–2186. [Google Scholar]
- Yuan, G.; Lin, H.; Wang, C.; Hong, K.; Liu, Y.; Li, J. 1h and 13c assignments of two new macrocyclic lactones isolated from streptomyces sp. 211726 and revised assignments of azalomycins f3a, f4aand f5a. Magn. Reson. Chem. 2011, 49, 30–37. [Google Scholar] [CrossRef]
- Fu, P.; Yang, C.; Wang, Y.; Liu, P.; Ma, Y.; Xu, L.; Su, M.; Hong, K.; Zhu, W. Streptocarbazoles a and b, two novel indolocarbazoles from the marine-derived actinomycete strain Streptomyces sp. Fma. Org. Lett. 2012, 14, 2422–2425. [Google Scholar] [CrossRef]
- Han, Y.; Tian, E.; Xu, D.; Ma, M.; Deng, Z.; Hong, K. Halichoblelide d, a new elaiophylin derivative with potent cytotoxic activity from mangrove-derived Streptomyces sp. 219807. Molecules 2016, 21, 970. [Google Scholar] [CrossRef] [Green Version]
- Voigt, W. Sulforhodamine b assay and chemosensitivity. In Chemosensitivity; Blumenthal, R.D., Ed.; Springer: Totowa, NJ, USA, 2005; pp. 39–48. [Google Scholar]
- Orellana, E.A.; Kasinski, A.L. Sulforhodamine b (srb) assay in cell culture to investigate cell proliferation. Bio-Protocol 2016, 6, e1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Chai, W.; Wang, W.; Song, T.; Lian, X.-Y.; Zhang, Z. Cytotoxic bagremycins from mangrove-derived streptomyces sp. Q22. J. Nat. Prod. 2017, 80, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Han, S.B.; Shin, Y.J.; Hyon, J.Y.; Wee, W.R. Cytotoxicity of voriconazole on cultured human corneal endothelial cells. Antimicrob. Agents Chemother. 2011, 55, 4519–4523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Liang, Z.; Chen, W.; Luo, X.; Fang, W.; Liao, S.; Lin, X.; Yang, B.; Wang, J.; Tang, L. Iakyricidins a–d, antiproliferative piericidin analogues bearing a carbonyl group or cyclic skeleton from Streptomyces iakyrus scsio ns104. J. Org. Chem 2019, 84, 12626–12631. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-L.; Jiang, S.-H.; Bu, X.-L.; Wang, J.-H.; Weng, J.-Y.; Yang, X.-M.; He, K.-Y.; Zhang, Z.-G.; Ao, P.; Xu, J. Structural diversity of anti-pancreatic cancer capsimycins identified in mangrove-derived Streptomyces xiamenensis 318 and post-modification via a novel cytochrome p450 monooxygenase. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the lactate dehydrogenase assay. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef]
- Baldwin, E.; Osheroff, N. Etoposide, topoisomerase ii and cancer. Curr. Med. Chem. Anticancer Agents 2005, 5, 363–372. [Google Scholar] [CrossRef]
- Giddings, L.-A.; Newman, D.J. Microbial natural products: Molecular blueprints for antitumor drugs. J. Ind. Microbiol. Biotechnol. 2013, 40, 1181–1210. [Google Scholar] [CrossRef]
- Singh, V.; Praveen, V.; Tripathi, D.; Haque, S.; Somvanshi, P.; Katti, S.; Tripathi, C. Isolation, characterization and antifungal docking studies of wortmannin isolated from Penicillium radicum. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef]
- Boehle, A.; Kurdow, R.; Boenicke, L.; Schniewind, B.; Faendrich, F.; Dohrmann, P.; Kalthoff, H. Wortmannin inhibits growth of human non-small-cell lung cancer in vitro and in vivo. Langenbecks Arch. Surg. 2002, 387, 234–239. [Google Scholar] [CrossRef]
- Kwon, H.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Marinomycins a− d, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J. Am. Chem. Soc. 2006, 128, 1622–1632. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.T.; Narender, T.; Kauffman, C.A.; Woolery, M.; Jensen, P.R.; Fenical, W. Saliniquinones a–f, new members of the highly cytotoxic anthraquinone-γ-pyrones from the marine actinomycete salinispora arenicola. Aust. J. Chem. 2010, 63, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Atta, H.M.; Ahmad, M.S. Antimycin-a antibiotic biosynthesis produced by streptomyces sp. Az-ar-262: Taxonomy, fermentation, purification and biological activities. Aust. J. Basic Appl. Sci. 2009, 3, 126–135. [Google Scholar]
- Shiomi, K.; Hatae, K.; Hatano, H.; Matsumoto, A.; Takahashi, Y.; Jiang, C.-L.; Tomoda, H.; Kobayashi, S.; Tanaka, H.; Ōmura, S. A new antibiotic, antimycin a 9, produced by Streptomyces sp. K01-0031. J. Antibiot. 2005, 58, 74–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.; Xu, Y.; McConnell, O.; Liu, L.; Li, Y.; Qi, S.; Huang, X.; Qian, P. Two antimycin a analogues from marine-derived actinomycete Streptomyces lusitanus. Mar. Drugs 2012, 10, 668–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.; Zhou, S.-W.; Chen, F.; Zheng, X.-H.; Shen, H.-F.; Lin, B.-R.; Zhou, G.-X. Neoantimycins a and b, two unusual benzamido nine-membered dilactones from marine-derived Streptomyces antibioticus h12-15. Molecules 2017, 22, 557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Yong, T.; Zhang, Y.; Su, J.; Jiao, C.; Xie, Y. Anti-tumor and anti-angiogenic ergosterols from Ganoderma lucidum. Front. Chem. 2017, 5, 85. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Fang, Y.; Zhang, M.; Lin, A.; Zhu, T.; Gu, Q.; Zhu, W. Six new ergosterols from the marine-derived fungus Rhizopus sp. Steroids 2008, 73, 19–26. [Google Scholar] [CrossRef]
- El-Gamal, A.A.; Wang, S.-K.; Dai, C.-F.; Duh, C.-Y. New nardosinanes and 19-oxygenated ergosterols from the soft coral Nephthea armata collected in taiwan. J. Nat. Prod. 2004, 67, 1455–1458. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Li, H.-Y.; Hu, C.; Sheng, H.-F.; Zhang, Y.; Lin, B.-R.; Zhou, G.-X. Ergosterols from the culture broth of marine Streptomyces anandii h41-59. Mar. Drugs 2016, 14, 84. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.; Wang, F.; Li, H.; Bao, Y.; Yang, Y.; Shen, H.; Lin, B.; Zhou, G. Secondary metabolites from marine-derived Streptomyces antibioticus strain h74-21. Nat. Prod. Res. 2016, 30, 2460–2467. [Google Scholar] [CrossRef] [PubMed]
- Mangamuri, U.; Muvva, V.; Poda, S.; Naragani, K.; Munaganti, R.K.; Chitturi, B.; Yenamandra, V. Bioactive metabolites produced by Streptomyces cheonanensis vuk-a from coringa mangrove sediments: Isolation, structure elucidation and bioactivity. 3 Biotech 2016, 6, 63. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, S.; Fredimoses, M.; Gnanadesigan, M. Anticancer property of sediment actinomycetes against mcf-7 and mda-mb-231 cell lines. Asian Pac. J. Trop. Biomed. 2012, 2, 92. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.T.-H.; Chan, C.-K.; Chan, K.-G.; Pusparajah, P.; Khan, T.M.; Ser, H.-L.; Lee, L.-H.; Goh, B.-H. Streptomyces sp. Mum256: A source for apoptosis inducing and cell cycle-arresting bioactive compounds against colon cancer cells. Cancers 2019, 11, 1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narendhran, S.; Rajiv, P.; Vanathi, P.; Sivaraj, R. Spectroscopic analysis of bioactive compounds from streptomyces cavouresis ku-v39: Evaluation of antioxidant and cytotoxicity activity. Int. J. Pharm. Pharm. Sci. 2014, 6, 319–322. [Google Scholar]
- Mithun, V.; Rao, C. Isolation and molecular characterization of anti-cancerous compound producing marine bacteria by using 16s rrna sequencing and gc-ms techniques. IJMER 2012, 2, 4510–4515. [Google Scholar]
- Gopi, M.; Dhayanithi, N.B.; Devi, K.N.; Kumar, T.T.A. Marine natural product, pyrrolo [-a] pyrazine–dione, hexahydro-(c7h10n2o2) of antioxidant properties from bacillus species at lakshadweep archipelago. J. Coast. Life Med. 2014, 2, 636–641. [Google Scholar]
- Brauns, S.C.; Milne, P.; Naude, R.; Van De Venter, M. Selected cyclic dipeptides inhibit cancer cell growth and induce apoptosis in ht-29 colon cancer cells. Anticancer Res. 2004, 24, 1713–1720. [Google Scholar]
- Cibi, R.; Nair, A.J. Purification of actinomycin d from Streptomyces parvulus isolated from mangrove ecosystem of kerala, india. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Sanjivkumar, M.; Babu, D.R.; Suganya, A.; Silambarasan, T.; Balagurunathan, R.; Immanuel, G. Investigation on pharmacological activities of secondary metabolite extracted from a mangrove associated actinobacterium Streptomyces olivaceus (msu3). Biocatal. Agric. Biotechnol. 2016, 6, 82–90. [Google Scholar] [CrossRef]
- Ser, H.-L.; Yin, W.-F.; Chan, K.-G.; Khan, T.M.; Goh, B.-H.; Lee, L.-H. Antioxidant and cytotoxic potentials of streptomyces gilvigriseus musc 26t isolated from mangrove soil in malaysia. Prog. Micobes Mol. Biol. 2018, 1, a0000002. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.-F.; Ser, H.-L.; Ab Mutalib, N.-S.; Saokaew, S.; Duangjai, A.; Khan, T.M.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. STREPTOMYCES monashensis sp. Nov., a novel mangrove soil actinobacterium from east malaysia with antioxidative potential. Sci. Rep. 2019, 9, 3056. [Google Scholar] [CrossRef] [PubMed]
- Spalding, M.; Kainuma, M.; Collins, L. World Atlas of Mangroves; Earthscan Routledge: New York, NY, USA, 2010. [Google Scholar]
- Rimando, A.M.; Olofsdotter, M.; Dayan, F.E.; Duke, S.O. Searching for rice allelochemicals: An example of bioassay-guided isolation. Agron. J. 2001, 93, 16–20. [Google Scholar] [CrossRef]
- Weller, M.G. A unifying review of bioassay-guided fractionation, effect-directed analysis and related techniques. Sensors 2012, 12, 9181–9209. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Handelsman, J. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 2003, 14, 303–310. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felczykowska, A.; Bloch, S.K.; Nejman-Faleńczyk, B.; Barańska, S. Metagenomic approach in the investigation of new bioactive compounds in the marine environment. Acta Biochim. Pol. 2012, 59. [Google Scholar] [CrossRef] [Green Version]
- Hugenholtz, P.; Tyson, G.W. Metagenomics. Nature 2008, 455, 481–483. [Google Scholar] [CrossRef]
- Kaur, R.; Rajesh, C.; Sharma, R.; Boparai, J.K.; Sharma, P.K. Metagenomic investigation of bacterial diversity of hot spring soil from manikaran, himachal pradesh, india. Ecol. Genet. Genom. 2018, 6, 16–21. [Google Scholar] [CrossRef]
- Wang, G.-Y.-S.; Graziani, E.; Waters, B.; Pan, W.; Li, X.; McDermott, J.; Meurer, G.; Saxena, G.; Andersen, R.J.; Davies, J. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2000, 2, 2401–2404. [Google Scholar] [CrossRef]
- Kleigrewe, K.; Almaliti, J.; Tian, I.Y.; Kinnel, R.B.; Korobeynikov, A.; Monroe, E.A.; Duggan, B.M.; Di Marzo, V.; Sherman, D.H.; Dorrestein, P.C. Combining mass spectrometric metabolic profiling with genomic analysis: A powerful approach for discovering natural products from cyanobacteria. J. Nat. Prod. 2015, 78, 1671–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herdini, C.; Mubarika, S.; Hariwiyanto, B.; Wijayanti, N.; Hosoyama, A.; Yamazoe, A.; Nojiri, H.; Widada, J. Secondary bioactive metabolite gene clusters identification of anticandida-producing Streptomyces sp. Gmr22 isolated from wanagama forest as revealed by genome mining approach. Indones. J. Pharm. 2017, 28, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Escribano, J.; Alt, S.; Bibb, M. Next generation sequencing of actinobacteria for the discovery of novel natural products. Mar. Drugs 2016, 14, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ser, H.-L.; Law, J.W.-F.; Tan, W.-S.; Yin, W.-F.; Chan, K.-G.; Lee, L.-H. Genome sequence of bioactive streptomycete isolated from mangrove forest in east malaysia, Streptomyces monashensis musc 1jt. Prog. Drug. Discov. Biomed. Sci. 2019, 2. [Google Scholar] [CrossRef]
- Musiol, R.; Serda, M.; Polanski, J. Prodrugs in photodynamic anticancer therapy. Curr. Pharm. Des. 2011, 17, 3548–3559. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Yoo, Y.J.; Lim, S.-K.; Cha, S.H.; Kim, J.-E.; Roe, J.-H.; Kim, J.F.; Yoon, Y.J. Complete genome sequence of Streptomyces venezuelae atcc 15439, a promising cell factory for production of secondary metabolites. J. Biotechnol. 2016, 219, 57–58. [Google Scholar] [CrossRef]
- Malla, S.; Niraula, N.P.; Liou, K.; Sohng, J.K. Improvement in doxorubicin productivity by overexpression of regulatory genes in Streptomyces peucetius. Res. Microbiol. 2010, 161, 109–117. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Van Lanen, S.G.; Baltz, R.H. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 2014, 41, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, H.A.; Low-Beinart, L.; Obiajulu, J.U.; Brady, S.F. Natural product discovery through improved functional metagenomics in streptomyces. J. Am. Chem. Soc. 2016, 138, 9341–9344. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Wang, B.; Zhao, H. Breaking the silence: New strategies for discovering novel natural products. Curr. Opin. Biotechnol. 2017, 48, 21–27. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Law, J.W.-F.; Law, L.N.-S.; Letchumanan, V.; Tan, L.T.-H.; Wong, S.H.; Chan, K.-G.; Ab Mutalib, N.-S.; Lee, L.-H. Anticancer Drug Discovery from Microbial Sources: The Unique Mangrove Streptomycetes. Molecules 2020, 25, 5365. https://doi.org/10.3390/molecules25225365
Law JW-F, Law LN-S, Letchumanan V, Tan LT-H, Wong SH, Chan K-G, Ab Mutalib N-S, Lee L-H. Anticancer Drug Discovery from Microbial Sources: The Unique Mangrove Streptomycetes. Molecules. 2020; 25(22):5365. https://doi.org/10.3390/molecules25225365
Chicago/Turabian StyleLaw, Jodi Woan-Fei, Lydia Ngiik-Shiew Law, Vengadesh Letchumanan, Loh Teng-Hern Tan, Sunny Hei Wong, Kok-Gan Chan, Nurul-Syakima Ab Mutalib, and Learn-Han Lee. 2020. "Anticancer Drug Discovery from Microbial Sources: The Unique Mangrove Streptomycetes" Molecules 25, no. 22: 5365. https://doi.org/10.3390/molecules25225365
APA StyleLaw, J. W. -F., Law, L. N. -S., Letchumanan, V., Tan, L. T. -H., Wong, S. H., Chan, K. -G., Ab Mutalib, N. -S., & Lee, L. -H. (2020). Anticancer Drug Discovery from Microbial Sources: The Unique Mangrove Streptomycetes. Molecules, 25(22), 5365. https://doi.org/10.3390/molecules25225365