Colombian Contributions Fighting Leishmaniasis: A Systematic Review on Antileishmanials Combined with Chemoinformatics Analysis
Abstract
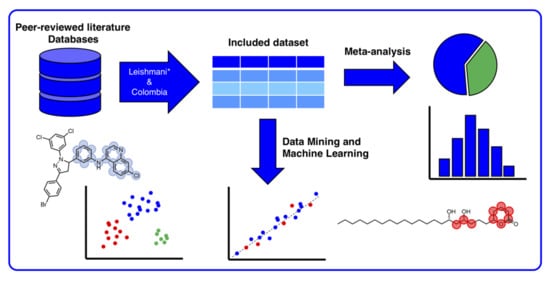
:1. Introduction
2. Results and Discussion
2.1. Study Characteristics
2.2. General Findings
2.3. Chemoinformatics Analyses on Retrieved Compounds
2.3.1. Chemical Space of Selected Compounds
2.3.2. Machine Learning
2.3.3. Drug-Likeness Filtering
3. Methods
3.1. Systematic Review
3.1.1. Search and Eligibility Criteria
3.1.2. Study Selection
3.1.3. Data Collection
3.2. Chemoinformatics Analysis
3.2.1. Data Preparation
3.2.2. Chemical Space by t-SNE
3.2.3. Random Forest
3.2.4. Support Vector Machines
3.2.5. Hyperparameter Optimization
3.2.6. Final Models
3.2.7. Analysis of Contributions by SHAP Values
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pan American Health Organization. Leishmaniases: Epidemiological Report in the Americas; Technical Report No. 8-2019; PAHO: Washington, DC, USA, 2019; pp. 1–10. [Google Scholar]
- Pan American Health Organization. Colombia: Cutaneous and Mucosal Leishmaniasis; PAHO: Washington, DC, USA, 2019; Volume 14, p. e0224351. [Google Scholar]
- Ramírez, J.D.; Hernández, C.; León, C.M.; Ayala, M.S.; Flórez, C.; González, C. Taxonomy, diversity, temporal and geographical distribution of Cutaneous Leishmaniasis in Colombia: A retrospective study. Sci. Rep. 2016, 6, 28266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montalvo, A.M.; Fraga, J.; Montano, I.; Monzote, L.; van der Auwera, G.; Marín, M.; Muskus, C. Identificación molecular de aislamientos clínicos de Leishmania spp. procedentes de Colombia con base en el gen hsp70. Biomédica 2016, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montalvo, A.M.; Fraga, J.; Tirado, D.; Blandón, G.; Alba, A.; van der Auwera, G.; Vélez, I.D.; Muskus, C. Detection and identification of Leishmania spp.: Application of two hsp70-based PCR-RFLP protocols to clinical samples from the New World. Parasitol. Res. 2017, 116, 1843–1848. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Almario, J.; Hernández, C.A.; Ovalle-Bracho, C. Geographical distribution of Leishmania species in Colombia, 1985–2017. Biomédica 2019, 39, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization. Manual of Procedures for Surveillance and Control of Leishmaniasis in the Americas; PAHO: Washington, DC, USA, 2019; ISBN 978-92-75-32063-1. [Google Scholar]
- An, I.; Harman, M.; Esen, M.; Celik, H. The effect of pentavalent antimonial compounds used in the treatment of cutaneous leishmaniasis on hemogram and biochemical parameters. Cutan. Ocul. Toxicol. 2019, 38, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Lyra, M.R.; Passos, S.R.L.; Pimentel, M.I.F.; Bedoya-Pacheco, S.J.; Valete-Rosalino, C.M.; Vasconcellos, E.C.F.; Antonio, L.F.; Saheki, M.N.; Salgueiro, M.M.; Santos, G.P.L.; et al. Pancreatic toxicity as an adverse effect induced by meglumine antimoniate therapy in a clinical trial for cutaneous leishmaniasis. Rev. Instit. Med. Trop. São Paulo 2016, 58. [Google Scholar] [CrossRef] [Green Version]
- Marques, S.A.; Merlotto, M.R.; Ramos, P.M.; Marques, S.A. American tegumentary leishmaniasis: Severe side effects of pentavalent antimonial in a patient with chronic renal failure. An. Bras. Dermatol. 2019, 94, 355–357. [Google Scholar] [CrossRef] [Green Version]
- Brito, N.C.; Rabello, A.; Cota, G. Efficacy of pentavalent antimoniate intralesional infiltration therapy for cutaneous leishmaniasis: A systematic review. PLoS ONE 2017, 12, e0184777. [Google Scholar] [CrossRef]
- Croft, S.L.; Sundar, S.; Fairlamb, A.H. Drug Resistance in Leishmaniasis. Clin. Microbiol. Rev. 2006, 19, 111–126. [Google Scholar] [CrossRef] [Green Version]
- Ponte-Sucre, A.; Gamarro, F.; Dujardin, J.-C.; Barrett, M.P.; López-Vélez, R.; García-Hernández, R.; Pountain, A.W.; Mwenechanya, R.; Papadopoulou, B. Drug resistance and treatment failure in leishmaniasis: A 21st century challenge. PLoS Negl. Trop. Dis. 2017, 11, e0006052. [Google Scholar] [CrossRef]
- Perilla-Gonzalez, Y.; Gomez-Suta, D.; Osorio, N.D.; Hurtado-Hurtado, N.; Baquero-Rodriguez, J.D.; Lopez-Isaza, A.F.; Lagos-Grisales, G.J.; Villegas, S.; Rodríguez-Morales, A.J. Study of the scientific production on leishmaniasis in Latin America. Recent Pat. Anti-Infect. Drug Discov. 2014, 9, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Mayorga, K.; Madariaga-Mazón, A.; Medina-Franco, J.L.M.; Maggiora, G. The impact of chemoinformatics on drug discovery in the pharmaceutical industry. Expert Opin. Drug Discov. 2020, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Cavasotto, C.N.; Aucar, M.G.; Adler, N.S. Computational chemistry in drug lead discovery and design. Int. J. Quantum Chem. 2019, 119, e25678. [Google Scholar] [CrossRef] [Green Version]
- Makhouri, F.R.; Ghasemi, J.B. Combating Diseases with Computational Strategies Used for Drug Design and Discovery. Curr. Top. Med. Chem. 2019, 18, 2743–2773. [Google Scholar] [CrossRef]
- Gillet, V.J. Applications of Chemoinformatics in Drug Discovery. In Biomolecular and Bioanalytical Techniques; Wiley: Hoboken, NJ, USA, 2019; pp. 17–36. [Google Scholar]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Lo, Y.-C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef]
- Réda, C.; Kaufmann, E.; Delahaye-Duriez, A. Machine learning applications in drug development. Comput. Struct. Biotechnol. J. 2020, 18, 241–252. [Google Scholar] [CrossRef]
- Halder, A.K.; Cordeiro, M.N.D.S. Advanced in Silico Methods for the Development of Anti- Leishmaniasis and Anti-Trypanosomiasis Agents. Curr. Med. Chem. 2020, 27, 697–718. [Google Scholar] [CrossRef]
- Scotti, L.; Ishiki, H.M.; Júnior, F.M.; da Silva, M.S.; Scotti, M. Artificial Neural Network Methods Applied to Drug Discovery for Neglected Diseases. Comb. Chem. High Throughput Screen. 2015, 18, 819–829. [Google Scholar] [CrossRef]
- Njogu, P.M.; Guantai, E.M.; Pavadai, E.; Chibale, K. Computer-Aided Drug Discovery Approaches against the Tropical Infectious Diseases Malaria, Tuberculosis, Trypanosomiasis, and Leishmaniasis. ACS Infect. Dis. 2016, 2, 8–31. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Andricopulo, A.D. Chemoinformatics Strategies for Leishmaniasis Drug Discovery. Front. Pharmacol. 2018, 9, 1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, J.D.; Tatonetti, N.P. Informatics and Computational Methods in Natural Product Drug Discovery: A Review and Perspectives. Front. Genet. 2019, 10, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, T. Harnessing the potential of natural products in drug discovery from a cheminformatics vantage point. Org. Biomol. Chem. 2017, 15, 9275–9282. [Google Scholar] [CrossRef] [PubMed]
- Olğaç, A.; Orhan, I.E.; Banoglu, E. The potential role ofin silicoapproaches to identify novel bioactive molecules from natural resources. Futur. Med. Chem. 2017, 9, 1663–1684. [Google Scholar] [CrossRef]
- Pereira, F.; Aires-De-Sousa, J. Computational Methodologies in the Exploration of Marine Natural Product Leads. Mar. Drugs 2018, 16, 236. [Google Scholar] [CrossRef] [Green Version]
- Herrera-Acevedo, C.; Scotti, L.; Alves, M.F.; Diniz, M.D.F.F.M.; Scotti, M.T. Computer-Aided Drug Design Using Sesquiterpene Lactones as Sources of New Structures with Potential Activity against Infectious Neglected Diseases. Molecules 2017, 22, 79. [Google Scholar] [CrossRef]
- Scotti, L.; Ishiki, H.; Mendonca, F.; Silva, M.S.; Scotti, M. In-silico Analyses of Natural Products on Leishmania Enzyme Targets. Mini-Rev. Med. Chem. 2015, 15, 253–269. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [Green Version]
- Trouiller, P.; Olliaro, P.; Torreele, E.; Orbinski, J.; Laing, R.; Ford, N. Drug development for neglected diseases: A deficient market and a public-health policy failure. Lancet 2002, 359, 2188–2194. [Google Scholar] [CrossRef] [Green Version]
- Murray, C.J.L.; Barber, R.M.; Foreman, K.J.; Ozgoren, A.A.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Abraham, J.P.; Abubakar, I.; Abu-Raddad, L.J.; et al. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990–2013: Quantifying the epidemiological transition. Lancet 2015, 386, 2145–2191. [Google Scholar] [CrossRef] [Green Version]
- Di Masi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef] [Green Version]
- Light, D.W.; Lexchin, J.R. Pharmaceutical research and development: What do we get for all that money? BMJ 2012, 345, e4348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surur, A.S.; Fekadu, A.; Makonnen, E.; Hailu, A. Challenges and Opportunities for Drug Discovery in Developing Countries: The Example of Cutaneous Leishmaniasis. ACS Med. Chem. Lett. 2020, 11, 2058–2062. [Google Scholar] [CrossRef] [PubMed]
- Alcântara, L.M.; Ferreira, T.C.; Gadelha, F.R.; Miguel, D.C. Challenges in drug discovery targeting TriTryp diseases with an emphasis on leishmaniasis. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional de Salud Estadísticas de Vigilancia Rutinaria. Available online: http://portalsivigila.ins.gov.co/VigilanciaRutinaria/rutinaria_2019.xlsx (accessed on 29 October 2020).
- Watts, K.R.; Tenney, K.; Crews, P. The structural diversity and promise of antiparasitic marine invertebrate-derived small molecules. Curr. Opin. Biotechnol. 2010, 21, 808–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, U.R.; Balasubramanian, S.; Oelschlaeger, T.A.; Grkovic, T.; Pham, N.B.; Quinn, R.J.; Hentschel, U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017, 17, e30–e41. [Google Scholar] [CrossRef]
- Imperatore, C.; Gimmelli, R.; Persico, M.; Casertano, M.; Guidi, A.; Saccoccia, F.; Ruberti, G.; Luciano, P.; Aiello, A.; Parapini, S.; et al. Investigating the Antiparasitic Potential of the Marine Sesquiterpene Avarone, Its Reduced Form Avarol, and the Novel Semisynthetic Thiazinoquinone Analogue Thiazoavarone. Mar. Drugs 2020, 18, 112. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.; Barreira, L.; Gangadhar, K.N.; Rodrigues, M.J.; Santos, T.; Varela, J.; Custódio, L. Natural products from marine invertebrates against Leishmania parasites: A comprehensive review. Phytochem. Rev. 2016, 15, 663–697. [Google Scholar] [CrossRef]
- Yamthe, L.R.T.; Appiah-Opong, R.; Fokou, P.V.T.; Nolé, T.; Boyom, F.F.; Nyarko, A.K.; Wilson, M. Marine Algae as Source of Novel Antileishmanial Drugs: A Review. Mar. Drugs 2017, 15, 323. [Google Scholar] [CrossRef] [Green Version]
- Álvarez-Bardón, M.; Pérez-Pertejo, Y.; Ordóñez, C.; Sepúlveda-Crespo, D.; Carballeira, N.M.; Tekwani, B.L.; Sankaranarayanan, M.; Martínez-Valladares, M.; García-Estrada, C.; Reguera, R.M.; et al. Screening Marine Natural Products for New Drug Leads against Trypanosomatids and Malaria. Mar. Drugs 2020, 18, 187. [Google Scholar] [CrossRef]
- Alzate, F.; Jimenez, N.; Weniger, B.; Bastida, J.; Gimenez, A. Antiprotozoal Activity of Ethanol Extracts of SomeBomareaSpecies. Pharm. Biol. 2008, 46, 575–578. [Google Scholar] [CrossRef] [Green Version]
- Weniger, B.; Robledo, S.; Arango, G.J.; Deharo, E.; Aragón, R.; Muñoz, V.; Callapa, J.; Lobstein, A.; Anton, R. Antiprotozoal activities of Colombian plants. J. Ethnopharmacol. 2001, 78, 193–200. [Google Scholar] [CrossRef]
- Lopez, R.; Cuca, L.; Delgado, G. Antileishmanial and immunomodulatory activity of Xylopia discreta. Parasite Immunol. 2009, 31, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Enciso, N.A.C.; Coy-Barrera, E.; Patiño, O.J.; Cuca, L.E.; Delgado, G. Evaluation of the Leishmanicidal Activity of Rutaceae and Lauraceae Ethanol Extracts on Golden Syrian Hamster (Mesocricetus auratus) Peritoneal Macrophages. Indian J. Pharm. Sci. 2014, 76, 188–197. [Google Scholar]
- Neira, L.F.; Mantilla, J.C.; Stashenko, E.; Escobar, P. Toxicidad, genotoxicidad y actividad anti-Leishmania de aceites esenciales obtenidos de cuatro (4) quimiotipos del género Lippia. Bol. Latinoam. Caribe Plantas Med. Aromat. 2018, 17, 68–83. [Google Scholar]
- Saez, J.; Granados, H.; Torres, B.; Velez, I.D.; Munoz, D. Leishmanicidal activity of Annona aff. spraguei seeds. Fitoterapia 1998, 69, 478–479. [Google Scholar]
- Jaramillo, M.; Arango, G.; González, M.; Robledo, S.; Velez, I. Cytotoxicity and antileishmanial activity of Annona muricata pericarp. Fitoterapia 2000, 71, 183–186. [Google Scholar] [CrossRef]
- Osorio, E.; Arango, G.J.; Jimenez, N.; Alzate, F.; Ruiz, G.; Gutiérrez, D.; Paco, M.A.; Giménez, A.; Robledo, S. Antiprotozoal and cytotoxic activities in vitro of Colombian Annonaceae. J. Ethnopharmacol. 2007, 111, 630–635. [Google Scholar] [CrossRef]
- Rodríguez, A.M.; Camargo, J.R.; García, F.J.B. Actividad in vitro de la mezcla de alcaloides de Ervatamia coronaria (Jacq) Staff. Apocynaceae sobre amastigotes de Leishmania braziliensis. Rev. Bras. Farm. 2008, 18, 350–355. [Google Scholar] [CrossRef]
- Arévalo, Y.; Robledo, S.; Muñoz, D.L.; Granados-Falla, D.; Cuca, L.E.; Delgado, G. Evaluación in vitro de la actividad de aceites esenciales de plantas colombianas sobre Leishmania Brazilien. Rev. Colomb. Cienc. Quim. Farm 2009, 38, 131–141. [Google Scholar]
- Céline, V.; Adriana, P.; Eric, D.; Joaquina, A.; Yannick, E.; Augusto, L.F.; Rosario, R.; Dionicia, G.; Michel, S.; Denis, C.; et al. Medicinal plants from the Yanesha (Peru): Evaluation of the leishmanicidal and antimalarial activity of selected extracts. J. Ethnopharmacol. 2009, 123, 413–422. [Google Scholar] [CrossRef]
- Calderon, A.I.; Romero, L.I.; Ortega-Barría, E.; Solis, P.N.; Zacchino, S.; Gimenez, A.; Pinzón, R.; Cáceres, A.; Tamayo, G.; Guerra, C.; et al. Screening of Latin American plants for antiparasitic activities against malaria, Chagas disease, and leishmaniasis. Pharm. Biol. 2010, 48, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Martínez, W.; Ospina, L.F.; Granados, D.; Delgado, G. In vitro studies on the relationship between the anti-inflammatory activity of Physalis peruviana extracts and the phagocytic process. Immunopharmacol. Immunotoxicol. 2009, 32, 63–73. [Google Scholar] [CrossRef]
- Sanchez-Suarez, J.; Riveros, I.; Delgado, G. Evaluation of the Leishmanicidal and Cytotoxic Potential of Essential Oils Derived from Ten Colombian Plants. Iran. J. Parasitol. 2013, 8, 129–136. [Google Scholar]
- Cardona Galeano, C.W.; Robledo-Restrepo, C.S.M.; Rojano, C.B.A.; Alzate-Guarin, C.F.; Muñoz-Herrera, D.L.; Saez-Vega, C.J. Leishmanicidal and antioxidant activity of extracts of Piper daniel-gonzalezii trel. (piperaceae). Rev. Cubana Plantas Med. 2013, 18, 268–277. [Google Scholar]
- Espitia-Baena, J.E.; Robledo-Restrepo, S.M.; Cuadrado-Cano, B.S.; Duran-Sandoval, H.R.; Gómez-Estrada, H.A. Perfil fitoquímico, actividad anti-Leishmania, hemolítica y toxicológica de Cordia dentata Poir. y Heliotropium indicum L. Rev. Cubana Plantas Med. 2014, 19, 208–224. [Google Scholar]
- Mesa, L.E.; Vasquez, D.; Lutgen, P.; Restrepo, A.M.; Robledo, S.; Velez, I.D.; Ortiz, I. In vitro and in vivo antileishmanial activity of Artemisia annua L. leaf powder and its potential usefulness in the treatment of uncomplicated cutaneous leishmaniasis in humans. Rev. Soc. Bras. Med. Trop. 2017, 50, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Marin, F.J.; Torres, O.L.; Robledo, S.; Doria, M.E. Estudio Fitoquímico y Evaluación de la Actividad Antioxidante y Leishmanicida de la Especie Pilocarpus alvaradoi (Rutaceae). Inform. Tecnol. 2018, 29, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Robledo, S.; Velez, I.D.; Schmidt, T.J. Arnica Tincture Cures Cutaneous Leishmaniasis in Golden Hamsters. Molecules 2018, 23, 150. [Google Scholar] [CrossRef] [Green Version]
- Laverde-Paz, M.J.; Echeverry, M.C.; Patarroyo, M.A.; Bello, F.J. Evaluating the anti-leishmania activity of Lucilia sericata and Sarconesiopsis magellanica blowfly larval excretions/secretions in an in vitro model. Acta Trop. 2018, 177, 44–50. [Google Scholar] [CrossRef]
- Patiño-Márquez, I.A.; Patiño-González, E.; Hernández-Villa, L.; Ortiz-Reyes, B.; Manrique-Moreno, M. Identification and evaluation of Galleria mellonella peptides with antileishmanial activity. Anal. Biochem. 2018, 546, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Vivero, R.J.; Mesa, G.B.; Robledo, S.M.; Herrera, C.X.M.; Cadavid-Restrepo, G. Enzymatic, antimicrobial, and leishmanicidal bioactivity of gram-negative bacteria strains from the midgut of Lutzomyia evansi, an insect vector of leishmaniasis in Colombia. Biotechnol. Rep. 2019, 24, e00379. [Google Scholar] [CrossRef] [PubMed]
- Patino, L.H.; Mendez, C.; Rodriguez, O.; Romero, Y.; Velandia, D.; Alvarado, M.; Pérez, J.; Duque, M.C.; Ramírez, J.D. Spatial distribution, Leishmania species and clinical traits of Cutaneous Leishmaniasis cases in the Colombian army. PLoS Negl. Trop. Dis. 2017, 11, e0005876. [Google Scholar] [CrossRef] [Green Version]
- Ovalle-Bracho, C.; Londoño-Barbosa, D.; Salgado-Almario, J.; González, C. Evaluating the spatial distribution of Leishmania parasites in Colombia from clinical samples and human isolates (1999 to 2016). PLoS ONE 2019, 14, e0214124. [Google Scholar] [CrossRef] [PubMed]
- Croft, S.L.; Yardley, V.; Kendrick, H. Drug sensitivity of Leishmania species: Some unresolved problems. Trans. R. Soc. Trop. Med. Hyg. 2002, 96, S127–S129. [Google Scholar] [CrossRef]
- Alcântara, L.M.; Ferreira, T.C.; Fontana, V.; Chatelain, E.; Moraes, C.B.; Freitas-Junior, L.H. A Multi-Species Phenotypic Screening Assay for Leishmaniasis Drug Discovery Shows That Active Compounds Display a High Degree of Species-Specificity. Molecules 2020, 25, 2551. [Google Scholar] [CrossRef]
- Dea-Ayuela, M.A.; Bilbao-Ramos, P.; Bolas-Fernández, F.; González, M.A. Synthesis and antileishmanial activity of C7- and C12-functionalized dehydroabietylamine derivatives. Eur. J. Med. Chem. 2016, 121, 445–450. [Google Scholar] [CrossRef]
- Emami, S.; Tavangar, P.; Keighobadi, M. An overview of azoles targeting sterol 14α-demethylase for antileishmanial therapy. Eur. J. Med. Chem. 2017, 135, 241–259. [Google Scholar] [CrossRef]
- Faiões, V.D.S.; da Frota, L.C.R.M.; Cunha-Júnior, E.F.; Barcellos, J.C.F.; da Silva, T.; Netto, C.D.; da Silva, S.A.G.; da Silva, A.J.M.; Costa, P.R.R.; Torres-Santos, E.C. Second-generation pterocarpanquinones: Synthesis and antileishmanial activity. J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 24, 35. [Google Scholar] [CrossRef] [Green Version]
- Fernández, O.L.; Diaz-Toro, Y.; Ovalle, C.; Valderrama, L.; Muvdi, S.; Rodríguez, I.; Gomez, M.A.; Saravia, N.G. Miltefosine and Antimonial Drug Susceptibility of Leishmania viannia Species and Populations in Regions of High Transmission in Colombia. PLoS Negl. Trop. Dis. 2014, 8, e2871. [Google Scholar] [CrossRef] [Green Version]
- Franco-Muñoz, C.; Manjarrés-Estremor, M.; Ovalle-Bracho, C. Intraspecies differences in natural susceptibility to amphotericine B of clinical isolates of Leishmania subgenus Viannia. PLoS ONE 2018, 13, e0196247. [Google Scholar] [CrossRef] [Green Version]
- Hefnawy, A.; Berg, M.; Dujardin, J.-C.; de Muylder, G. Exploiting Knowledge on Leishmania Drug Resistance to Support the Quest for New Drugs. Trends Parasitol. 2017, 33, 162–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, D.L.; Robledo, S.M.; Kolli, B.K.; Dutta, S.; Chang, K.P.; Muskus, C. Leishmania (Viannia) panamensis: An in vitro assay using the expression of GFP for screening of antileishmanial drug. Exp. Parasitol. 2009, 122, 134–139. [Google Scholar] [CrossRef] [Green Version]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Katsuno, K.; Burrows, J.N.; Duncan, K.; van Huijsduijnen, R.H.; Kaneko, T.; Kita, K.; Mowbray, C.E.; Schmatz, D.; Warner, P.; Slingsby, B.T. Hit and lead criteria in drug discovery for infectious diseases of the developing world. Nat. Rev. Drug Discov. 2015, 14, 751–758. [Google Scholar] [CrossRef]
- MACCS Structural Keys; Accelrys: San Diego, CA, USA, 2011.
- Morgan, H.L. The Generation of a Unique Machine Description for Chemical Structures-A Technique Developed at Chemical Abstracts Service. J. Chem. Doc. 1965, 5, 107–113. [Google Scholar] [CrossRef]
- RDKit: Cheminformatics and Machine Learning Software. Available online: http://rdkit.org (accessed on 21 May 2020).
- Van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Brown, S.P.; Muchmore, S.W.; Hajduk, P.J. Healthy skepticism: Assessing realistic model performance. Drug Discov. Today 2009, 14, 420–427. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Systems (NIPS 2017), Long Beach, California, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 4768–4777, ISBN 978-1-5108-6096-4. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; Degrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Rodríguez-Pérez, R.; Bajorath, J. Interpretation of Compound Activity Predictions from Complex Machine Learning Models Using Local Approximations and Shapley Values. J. Med. Chem. 2019, 63, 8761–8777. [Google Scholar] [CrossRef]
- Rodríguez-Pérez, R.; Bajorath, J. Interpretation of machine learning models using shapley values: Application to compound potency and multi-target activity predictions. J. Comput. Mol. Des. 2020, 34, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staderini, M.; Piquero, M.; Abengózar, M.Á.; Nachér-Vázquez, M.; Romanelli, G.; López-Alvarado, P.; Rivas, L.; Bolognesi, M.L.; Menéndez, J.C. Structure-activity relationships and mechanistic studies of novel mitochondria-targeted, leishmanicidal derivatives of the 4-aminostyrylquinoline scaffold. Eur. J. Med. Chem. 2019, 171, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Fakhfakh, M.A.; Fournet, A.; Prina, E.; Mouscadet, J.F.; Franck, X.; Hocquemiller, R.; Figadère, B. Synthesis and biological evaluation of substituted quinolines: Potential treatment of protozoal and retroviral co-infections. Bioorg. Med. Chem. 2003, 11, 5013–5023. [Google Scholar] [CrossRef]
- Mateen, F.J.; Oh, J.; Tergas, A.I.; Bhayani, N.H.; Kamdar, B.B. Titles versus titles and abstracts for initial screening of articles for systematic reviews. Clin. Epidemiol. 2013, 5, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- López-López, E.; Naveja, J.J.; Medina-Franco, J.L. DataWarrior: An evaluation of the open-source drug discovery tool. Expert Opin. Drug Discov. 2019, 14, 335–341. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, O.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Fawagreh, K.; Gaber, M.M.; Elyan, E. Random forests: From early developments to recent advancements. Syst. Sci. Control. Eng. 2014, 2, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Nembrini, S.; König, I.R.; Wright, M.N. The revival of the Gini importance? Bioinformatics 2018, 34, 3711–3718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Shapley, L.S. A value for n-person games. In Contributions to the Theory of Games; Kuhn, H.W., Tucker, A.W., Eds.; Princeton University Press: Princenton, NJ, USA, 1953; Volume 2, pp. 307–317. [Google Scholar]
- Lundberg, S.M. A Game Theoretic Approach to Explain the Output of Any Machine Learning Model. Available online: https://github.com/slundberg/shap#citations (accessed on 29 October 2020).










| Leishmania Species a | Parasites Form b | EC50 (μg/mL) c | E/F d | Source e | Ref h |
|---|---|---|---|---|---|
| L. braziliensis; L. infantum; L. panamensis | Promastigote | N/A e | 1 | Annona spraguei | [51] |
| L. braziliensis; L. panamensis | Promastigote | N/A e | 3 | Annona muricata | [52] |
| L. amazonensis; L. braziliensis; L. infantum; L. panamensis | Promastigote, Intracellular amastigote | 1.30 | 88 | Conobea scoparioides | [47] |
| L. amazonensis; L. braziliensis; L. donovani | Promastigote | 10.70 | 36 | Rollinia pittieri | [53] |
| L. amazonensis; L. braziliensis; L. donovani | Promastigote | 4.90 | 26 | Bomarea setacea | [46] |
| L. braziliensis | Intracellular amastigote | 12.40 | 1 | Ervatamia coronaria | [54] |
| L. panamensis | Intracellular amastigote | 6.25 | 8 | Xylopia discreta | [48] |
| L. braziliensis | Promastigote | 17.40 | 13 | Rosmarinus officinalis | [55] |
| L. amazonensis | Axenic amastigotes | 9.00 | 94 | Renealmia alpinia | [56] |
| L. mexicana | Axenic amastigotes | >50.00 | 452 f | Several g | [57] |
| L. panamensis | Intracellular amastigote | 15.40 | 6 | Physalis peruviana | [58] |
| L. panamensis; L. braziliensis; L. major; L. guyanensis | Promastigote | 42.23 | 10 | Origanum vulgare | [59] |
| L. panamensis | Intracellular amastigote, Axenic amastigotes | 38.50 | 6 | Piper daniel-gonzalezii | [60] |
| L. panamensis | Intracellular amastigote | 18.50 | 13 | Heliotropium indicum | [61] |
| L. panamensis; L. major | Promastigote, Intracellular amastigote | 6.16 | 3 | Zanthoxyllum monophyllum | [49] |
| L. panamensis | Intracellular amastigote | 48.07 | 1 | Artemisia annua | [62] |
| L. braziliensis; L. panamensis | Promastigote, Intracellular amastigote | 9.19 | 4 | Lippia alba | [50] |
| L. panamensis | Intracellular amastigote | 30.70 | 8 | Pilocarpus alvaradoi | [63] |
| L. braziliensis | in vivo on golden hamsters | N/A f | 1 | Arnica montana | [64] |
| L. panamensis | Promastigote, Intracellular amastigote | 23.42 | 2 | Sarconesiopsis magellanica | [65] |
| L. panamensis | Promastigote | N/A f | 4 | Galleria mellonella | [66] |
| L. infantum; L. braziliensis | Promastigote | 47.70 | 12 | Enterobacter hormaechei | [67] |
| Parasite Form a | Antileishmanial Activity Category b | Number of Records c |
|---|---|---|
| Intracellular amastigotes | High | 127 |
| Intermediate | 68 | |
| Low | 221 | |
| Not Determined | 162 | |
| Not Available | 112 | |
| Axenic amastigotes | High | 28 |
| Intermediate | 27 | |
| Low | 37 | |
| Not Determined | 32 | |
| Not Available | 28 | |
| Promastigotes | High | 29 |
| Intermediate | 44 | |
| Low | 81 | |
| Not Determined | 50 | |
| Not Available | 14 | |
| Total d | 1060 | |
| Validation Parameter a | M1 b,d | M2 b,e | M3 c,d | M4 c,e |
|---|---|---|---|---|
| R2train | 0.813 | 0.925 | 0.847 | 0.849 |
| MAEtrain | 0.250 | 0.155 | 0.202 | 0.183 |
| R2CV | 0.621 | 0.621 | 0.600 | 0.592 |
| MAECV | 0.359 | 0.354 | 0.370 | 0.358 |
| R2test | 0.670 | 0.666 | 0.583 | 0.689 |
| MAEtest | 0.322 | 0.324 | 0.352 | 0.339 |
| Compound | Species | EC50 a | Cellular Line | IC50 b | SI c | Ro5 d Violations |
|---|---|---|---|---|---|---|
| 3 | L. panamensis | 4.03 | U-937 | 15.6 | 3.9 | 0 |
| 84 | L. braziliensis | 2.26 | U-937 | 6.99 | 3.1 | 1 |
| 85 | L. braziliensis | 2.53 | U-937 | 8.75 | 3.5 | 2 |
| 191 | L. panamensis | 0.57 | U-937 | 8.02 | 14 | 0 |
| 192 | L. panamensis | 7.48 | U-937 | 17.7 | 2.4 | 0 |
| 341 | L. panamensis | 4.81 | U-937 | 12.7 | 2.6 | 0 |
| 343 | L. panamensis | 6.18 | U-937 | 21.3 | 3.4 | 0 |
| 345 | L. panamensis | 5.90 | U-937 | 15.6 | 2.6 | 0 |
| 465 | L. braziliensis | 3.30 | BMDM | 724 | 91 | 1 |
| 487 | L. panamensis | 7.07 | U-937 | 19.7 | 2.8 | 2 |
| 489 | L. panamensis | 3.70 | U-937 | 9.62 | 2.6 | 2 |
| 490 | L. panamensis | 3.41 | U-937 | 9.69 | 2.8 | 1 |
| 499 | L. panamensis | 6.50 | U-937 | 16.7 | 2.6 | 2 |
| 511 | L. panamensis | 4.47 | U-937 | 322 | 72 | 2 |
| 566 | L. panamensis | 5.51 | U-937 | 13.1 | 2.4 | 1 |
| 804 | L. panamensis | 0.60 | U-937 | 3.87 | 3.9 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Suárez, J.; Bernal, F.A.; Coy-Barrera, E. Colombian Contributions Fighting Leishmaniasis: A Systematic Review on Antileishmanials Combined with Chemoinformatics Analysis. Molecules 2020, 25, 5704. https://doi.org/10.3390/molecules25235704
Sánchez-Suárez J, Bernal FA, Coy-Barrera E. Colombian Contributions Fighting Leishmaniasis: A Systematic Review on Antileishmanials Combined with Chemoinformatics Analysis. Molecules. 2020; 25(23):5704. https://doi.org/10.3390/molecules25235704
Chicago/Turabian StyleSánchez-Suárez, Jeysson, Freddy A. Bernal, and Ericsson Coy-Barrera. 2020. "Colombian Contributions Fighting Leishmaniasis: A Systematic Review on Antileishmanials Combined with Chemoinformatics Analysis" Molecules 25, no. 23: 5704. https://doi.org/10.3390/molecules25235704
APA StyleSánchez-Suárez, J., Bernal, F. A., & Coy-Barrera, E. (2020). Colombian Contributions Fighting Leishmaniasis: A Systematic Review on Antileishmanials Combined with Chemoinformatics Analysis. Molecules, 25(23), 5704. https://doi.org/10.3390/molecules25235704