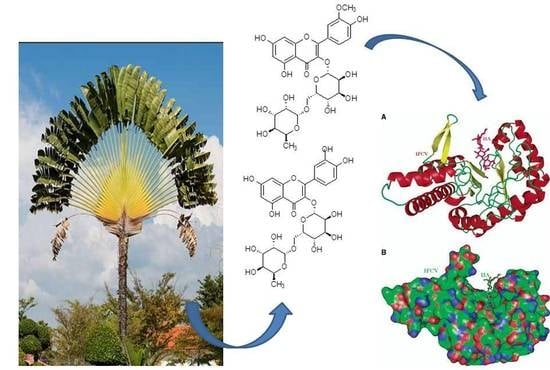
Bioassay-Guided Isolation, Metabolic Profiling, and Docking Studies of Hyaluronidase Inhibitors from Ravenala madagascariensis
Abstract
:1. Introduction
2. Results
2.1. Metabolic Profiling
2.2. Identification of Purified Compounds
2.3. Modeling Study Exhibiting the Binding Ability of Polyphenolic Compounds to HAase
3. Materials and Methods
3.1. Plant Material
3.2. Chemicals and Reagents
3.3. Extraction and Isolation
3.4. Biological Activity Study
3.4.1. Preparation of Hyaluronidase
3.4.2. Hyaluronidase Inhibition Assay
3.5. Metabolic Profiling
3.6. Molecular Docking Investigation
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kendall, A.C.; Nicolaou, A. Bioactive lipid mediators in skin inflammation and immunity. Prog. Lipid Res. 2013, 52, 141–164. [Google Scholar] [CrossRef] [PubMed]
- Farris, P.K. Innovative cosmeceuticals: Sirtuin activators and anti-glycation compounds. Semin. Cutan. Med. Surg. 2011, 30, 163. [Google Scholar] [CrossRef] [PubMed]
- Tzellos, T.; Klagas, I.; Vahtsevanos, K.; Triaridis, S.; Printza, A.; Kyrgidis, A.; Karakiulakis, G.; Zouboulis, C.; Papakonstantinou, E. Extrinsic ageing in the human skin is associated with alterations in the expression of hyaluronic acid and its metabolizing enzymes. Exp. Dermatol. 2009, 18, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- An, B.J.; Kwak, J.H.; Park, J.M.; Lee, J.Y.; Park, T.S.; Lee, J.T.; Son, J.H.; Jo, C.; Byun, M.W. Inhibition of enzyme activities and the antiwrinkle effect of polyphenol isolated from the persimmon leaf (Diospyros kaki folium) on human skin. Dermatol. Surg. 2005, 31, 848–855. [Google Scholar] [CrossRef]
- Papakonstantinou, E.; Roth, M.; Karakiulakis, G. Hyaluronic acid: A key molecule in skin aging. Derm. Endocrinol. 2012, 4, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Taofiq, O.; González-Paramás, A.M.; Martins, A.; Barreiro, M.F.; Ferreira, I.C. Mushrooms extracts and compounds in cosmetics, cosmeceuticals and nutricosmetics—A review. Ind. Crop. Prod. 2016, 90, 38–48. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Bharadwaj, A.G.; Casper, A.; Barkley, J.; Barycki, J.J.; Simpson, M.A. Hyaluronidase activity of human Hyal1 requires active site acidic and tyrosine residues. J. Biol. Chem. 2009, 284, 9433–9442. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, N.; Thomas, N.; Sivasothy, Y.; Lee, V.; Liew, S.; Noorbatcha, I.; Awang, K. Hyaluronidase inhibitory activity of pentacylic triterpenoids from Prismatomeris tetrandra (Roxb.) K. Schum: Isolation, synthesis and QSAR study. Int. J. Mol. Sci. 2016, 17, 143. [Google Scholar] [CrossRef]
- Girish, K.; Kemparaju, K.; Nagaraju, S.; Vishwanath, B. Hyaluronidase inhibitors: A biological and therapeutic perspective. Curr. Med. Chem. 2009, 16, 2261–2288. [Google Scholar] [CrossRef]
- Liyanaarachchi, G.D.; Samarasekera, J.K.R.R.; Mahanama, K.R.R.; Hemalal, K.D.P. Tyrosinase, elastase, hyaluronidase, inhibitory and antioxidant activity of Sri Lankan medicinal plants for novel cosmeceuticals. Ind. Crops Prod. 2018, 111, 597–605. [Google Scholar] [CrossRef]
- Jegasothy, S.M.; Zabolotniaia, V.; Bielfeldt, S. Efficacy of a new topical nano-hyaluronic acid in humans. J. Clin. Aesthetic Dermatol. 2014, 7, 27. [Google Scholar]
- Necas, J.; Bartosikova, L.; Brauner, P.; Kolar, J. Hyaluronic acid (hyaluronan): A review. Vet. Med. 2008, 53, 397–411. [Google Scholar] [CrossRef] [Green Version]
- Daniel, Z.; Marta, O.; Agnieszka, G.; Robert, V.; Rafal, K.; Bogucka-Kocka, A. Phytochemical content and pharma-nutrition study on Eleutherococcus senticosus fruits intractum. Oxidative Med. Cell. Longev. 2016, 2016, 9270691. [Google Scholar]
- Daniel, Z.; Marta, O.; Kuz′niewski, R.; Verpoorte, R.; Nowak, R.; Helena, D. Smolarz: LC-ESI-MS/MS profiling of phenolics from Eleutherococcus spp. inflorescences, structure-activity relationship as antioxidants, inhibitors of hyaluronidase and acetylcholinesterase. Saudi Pharm. J. 2017, 25, 734–743. [Google Scholar]
- Scotti, L.; Kumar Singla, R.; Mitsugu Ishiki, H.; Mendonca, J.B.; Sobral da Silva, M.; Barbosa Filho, M.; Tullius Scotti, M. Recent advancement in natural hyaluronidase inhibitors. Curr. Top. Med. Chem. 2016, 16, 2525–2531. [Google Scholar] [CrossRef]
- Murata, T.; Miyase, T.; Yoshizaki, F. Hyaluronidase inhibitors from Keiskea japonica. Chem. Pharm. Bull. 2012, 60, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.-J.; Yang, R.; You, J.; Qu, L.-B.; Sun, Y.-J. Spectroscopic and Docking Studies on the Binding of Liquiritigenin with Hyaluronidase for Antiallergic Mechanism. Scientifica (Cairo) 2016, 2016, 9178097. [Google Scholar] [CrossRef] [Green Version]
- Bose, B.; Choudhury, H.; Tandon, P.; Kumaria, S. Studies on secondary metabolite profiling, anti-inflammatory potential, in vitro photoprotective and skin-aging related enzyme inhibitory activities of Malaxis acuminata, a threatened orchid of nutraceutical importance. J. Photochem. Photobiol. B Biol. 2017, 173, 686–695. [Google Scholar] [CrossRef]
- Xu, M.; Zhu, H.-T.; Cheng, R.-R.; Wang, D.; Yang, C.-R.; Tanaka, T.; Kouno, I.; Zhang, Y.-J. Antioxidant and hyaluronidase inhibitory activities of diverse phenolics in Phyllanthus emblica. Nat. Prod. Res. 2016, 30, 2726–2729. [Google Scholar] [CrossRef]
- Perera, H.D.S.M.; Samarasekera, J.K.R.R.; Handunnetti, S.M.; Weerasena, O.V.D.S.J. In vitro anti-inflammatory and anti-oxidant activities of Sri Lankan medicinal plants. Ind. Crop. Prod. 2016, 94, 610–620. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Maity, N.; Nema, N.K.; Sarkar, B.K. Bioactive compounds from natural resources against skin aging. Phytomedicine 2011, 19, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Tomohara, K.; Ito, T.; Onikata, S.; Kato, A.; Adachi, I. Discovery of hyaluronidase inhibitors from natural products and their mechanistic characterization under DMSO-perturbed assay conditions. Bioorganic Med. Chem. Lett. 2017, 27, 1620–1623. [Google Scholar] [CrossRef] [PubMed]
- Onifade, A.; Bello, M.; Fadipe, D. Bioassay directed fractionation of antibacterial compounds from traveller’s tree (Ravenala madagascariensis sonnerat) and its phytochemical constituents. Int. J. Bioassays 2015, 4, 4299–4304. [Google Scholar]
- Ramiarantsoa, H.; Yao-Kouassi, P.; Kanko, C.; Assi, K.; Djakoure, A.; Tonzibo, F. Chemical constituents of the antidiabetic plant Ravenala madagascariensis. Int. J. Pharm. Sci. Res. 2014, 5, 5503. [Google Scholar]
- Reyad-Ul-Ferdous, M.; Nasir, U.; Shahjahan, D.; Mosharaf, H.; Arman, M.; Ariful, I. Preliminary in-vitro potential phytochemicals investigation of barks of Ravenala madagascariensis Sonnerat. World J. Pharm. Pharm. Sci. (WJPPS) 2014, 3, 1418–1423. [Google Scholar]
- Renimel, I.; Andre, P. Extract of the Plant Ravenala Madagascariensis and USE as Cosmetic Hydrating Agent. U.S. Patent 8,697,153, 15 April 2014. [Google Scholar]
- Dehaghani, Z.A.; Asghari, G.; Dinani, M.S. Isolation and Identification of Nicotiflorin and Narcissin from the Aerial Parts of Peucedanum aucheri Boiss. J. Agric. Sci. Technol. A 2017, 7, 45–51. [Google Scholar]
- Sikorska, M.; Matławska, I. Kaempferol, isorhamnetin and their glycosides in the flowers of Asclepias syriaca L. Acta Pol. Pharm. 2001, 58, 269–272. [Google Scholar]
- Holscher, D.; Schneider, B. Phenalenones from Strelitzia reginae. J. Nat. Prod. 2000, 63, 1027–1028. [Google Scholar] [CrossRef]
- Nowak, S.; Wolbis, M. Flavonoids from some species of genus Scopolia Jacq. Acta Pol. Pharm. 2002, 59, 275–280. [Google Scholar]
- Mervoyer, C.; Portet, B.; Giboulot, J.; Lubrano, C. Characterization of flavonoid glycosides in traveller′s tree (Ravenala madagascariensis S.) leaves by HPLC-DAD-ESI-MSn. Planta Med. 2012, 78, PJ40. [Google Scholar] [CrossRef]
- Liu, H.; Mou, Y.; Zhao, J.; Wang, J.; Zhou, L.; Wang, M.; Wang, D.; Han, J.; Yu, Z.; Yang, F. Flavonoids from Halostachys caspica and their antimicrobial and antioxidant activities. Molecules 2010, 15, 7933. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, S.M.; Motaal, A.A. Isolation of new cytotoxic metabolites from Cleome droserifolia growing in Egypt. Z. Nat. C 2012, 67, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Budzianowski, J. Kaempferol glycosides from Hosta ventricosa. Phytochemistry 1990, 29, 3643–3647. [Google Scholar] [CrossRef]
- Li, Y.-L.; Li, J.; Wang, N.-L.; Yao, X.-S. Flavonoids and a new polyacetylene from Bidens parviflora Willd. Molecules 2008, 13, 1931. [Google Scholar] [CrossRef] [PubMed]
- Kpegba, K.; Agbonon, A.; Petrovic, A.G.; Amouzou, E.; Gbeassor, M.; Proni, G.; Nesnas, N. Epiafzelechin from the root bark of cassia sieberiana: Detection by DART mass spectrometry, spectroscopic characterization, and antioxidant properties. J. Nat. Prod. 2010, 74, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Donovan, J.L.; Luthria, D.L.; Stremple, P.; Waterhouse, A.L. Analysis of (+)-catechin,(−)-epicatechin and their 3′-and 4′-O-methylated analogs: A comparison of sensitive methods. J. Chromatogr. B Biomed. Sci. Appl. 1999, 726, 277–283. [Google Scholar] [CrossRef]
- Usman, A.; Thoss, V.; Nur-e-Alam, M. Isolation of (-)-Epicatechin from Trichilia emetica Whole Seeds. Am. J. Org. Chem. 2016, 6, 81–85. [Google Scholar]
- Stern, R.; Jedrzejas, M.J. Hyaluronidases: Their genomics, structures, and mechanisms of action. Chem. Rev. 2006, 106, 818–839. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.F.; Willett, N.P. Rapid plate method for screening hyaluronidase and chondroitin sulfatase-producing microorganisms. Appl. Microbiol. 1968, 16, 1434. [Google Scholar] [CrossRef] [Green Version]
- Mc Cook, J.P.; Dorogi, P.L.; Vasily, D.B.; Cefalo, R.D. In vitro inhibition of hyaluronidase by sodium copper chlorophyllin complex and chlorophyllin analogs. Clin. Cosmet. Investig. Derm. 2015, 8, 443–448. [Google Scholar]
- Sumantran, V.N.K.A.; Harsulkar, A. Hyaluronidase and collagenase inhibitory activities of the herbal formulation Triphala guggulu. J. Biosci. 2007, 4, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Satardekar, K.V.D.M. Anti-ageing ability of Terminalia species with special reference to hyaluronidase, elastase inhibition and collagen synthesis in vitro. Int. J. Pharmacogn. Phytochem. Res. 2010, 2, 30–34. [Google Scholar]
- Abdelmohsen, U.R.; Cheng, C.; Viegelmann, C.; Zhang, T.; Grkovic, T.; Ahmed, S.; Quinn, R.J.; Hentschel, U.; Edrada-Ebel, R. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49. Mar. Drugs 2014, 12, 1220–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelhafez, O.H.; Fawzy, M.A.; Fahim, J.R.; Desoukey, S.Y.; Krischke, M.; Mueller, M.J.; Abdelmohsen, U.R. Hepatoprotective potential of Malvaviscus arboreus against carbon tetrachloride-induced liver injury in rats. PLoS ONE 2018, 13, e0202362. [Google Scholar] [CrossRef]
- Boström, J.; Greenwood, J.R.; Gottfries, J. Assessing the performance of OMEGA with respect to retrieving bioactive conformations. J. Mol. Graph. Model. 2003, 21, 449–462. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |




| Crude Extracts | Hyaluronidase Inhibition | Leaf Partitions | Hyaluronidase Inhibition | Pure Compounds | Hyaluronidase Inhibition |
|---|---|---|---|---|---|
| Leaves | 78.9% ± 0.023 | n-Hexane | 28.6% ± 0.065 | Narcissin | 27.2% ± 0.043 |
| Stem | 56.1% ± 1.082 | DCM | 3.8% ± 1.044 | Rutin | 23.7% ± 01.070 |
| Flower | 9.4% ± 0.1012 | EtOAc | 32% ± 0.1334 | Epicatechin | 34.4% ± 0.038 |
| Root | 20.5% ± 0.006 | n-BuOH | 64.3% ± 0.015 | Epiafzelechin | 36.5% ± 0.045 |
| Tannic acid | 95.4% ± 0.006 | total 70% EtOH | 90 % ± 0.006 | Isorhamnetin 7-O-glucoside | 11.4% ± 0.022 |
| Kaempferol | 8.7% ± 0.039 |
| Compound | Score | Average Number of Poses Per Run |
|---|---|---|
| Narcissin | −6.853 | 10 |
| Quercetin-3-O-glucoside | −6.088 | 10 |
| Rutin | −7.119 | 10 |
| 8-hydroxy-7-methoxy-6-phenylphenalen-1-one | −4.54 | 10 |
| Hydroxyanigorufone | −4.306 | 7 |
| Epiafzelechin | −4.436 | 8 |
| Epicatechin | −4.852 | 8 |
| Lachnanthocarpone | −4.539 | 6 |
| Confertoside | −5.936 | 10 |
| Dendroside D | −6.701 | 2 |
| Loliolide | −4.021 | 8 |
| Shiraiachrome A | −6.08 | 9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
M. Mohamed, E.; H. Hetta, M.; Rateb, M.E.; A. Selim, M.; M. AboulMagd, A.; A. Badria, F.; Abdelmohsen, U.R.; Alhadrami, H.A.; M. Hassan, H. Bioassay-Guided Isolation, Metabolic Profiling, and Docking Studies of Hyaluronidase Inhibitors from Ravenala madagascariensis. Molecules 2020, 25, 1714. https://doi.org/10.3390/molecules25071714
M. Mohamed E, H. Hetta M, Rateb ME, A. Selim M, M. AboulMagd A, A. Badria F, Abdelmohsen UR, Alhadrami HA, M. Hassan H. Bioassay-Guided Isolation, Metabolic Profiling, and Docking Studies of Hyaluronidase Inhibitors from Ravenala madagascariensis. Molecules. 2020; 25(7):1714. https://doi.org/10.3390/molecules25071714
Chicago/Turabian StyleM. Mohamed, Esraa, Mona H. Hetta, Mostafa E. Rateb, Mohamed A. Selim, Asmaa M. AboulMagd, Farid A. Badria, Usama Ramadan Abdelmohsen, Hani A. Alhadrami, and Hossam M. Hassan. 2020. "Bioassay-Guided Isolation, Metabolic Profiling, and Docking Studies of Hyaluronidase Inhibitors from Ravenala madagascariensis" Molecules 25, no. 7: 1714. https://doi.org/10.3390/molecules25071714
APA StyleM. Mohamed, E., H. Hetta, M., Rateb, M. E., A. Selim, M., M. AboulMagd, A., A. Badria, F., Abdelmohsen, U. R., Alhadrami, H. A., & M. Hassan, H. (2020). Bioassay-Guided Isolation, Metabolic Profiling, and Docking Studies of Hyaluronidase Inhibitors from Ravenala madagascariensis. Molecules, 25(7), 1714. https://doi.org/10.3390/molecules25071714