Development of a Wine Metabolomics Approach for the Authenticity Assessment of Selected Greek Red Wines
Abstract
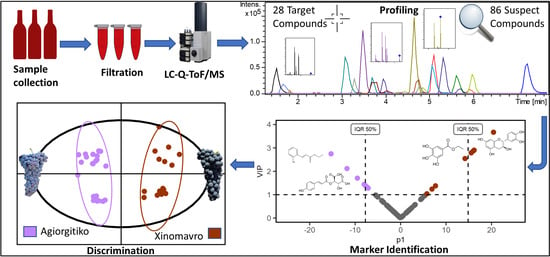
:1. Introduction
2. Results and Discussion
2.1. Method Validation Results
2.2. Target Screening
2.3. Suspect Screening Results
2.4. Statistical Analysis
2.4.1. Data Processing
2.4.2. Chemometrics
2.4.3. Identification of Markers
3. Materials and Methods
3.1. Sample Collections
3.2. Chemicals and Reagents
3.3. Preparation of Standard Stock Solutions
3.4. Sample Preparation
3.5. UHPLC–QToF-MS Analysis
3.6. Method Validation
3.7. Screening Strategies
3.7.1. Target Screening
3.7.2. Suspect Screening Strategy
3.8. Data Processing and Chemometrics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Valamoti, S.M.; Mangafa, M.; Koukouli-Chrysanthaki, C.; Malamidou, D. Grape-pressings from northern Greece: The earliest wine in the Aegean? Antiquity 2007, 81, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Hope, K. Wine in Greece: Rediscovered Country. Available online: https://www.ft.com/content/cbbefe38-4a27-11e9-bde6-79eaea5acb64 (accessed on 10 April 2021).
- Holmberg, L. Wine fraud. Int. J. Wine Res. 2010, 2, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, A.; Countryman, A.M. In Vino ‘No’ Veritas: Impacts of fraud on wine imports in China. Aust. J. Agric. Resour. Econ. 2019, 63, 742–758. [Google Scholar] [CrossRef] [Green Version]
- Fougere, E.; Kaplan, E.K.; Collins, C.A. Pricing uncertainty in wine markets following the Rudy Kurniawan scandal. J. Wine Res. 2020, 31, 1–5. [Google Scholar] [CrossRef]
- Versari, A.; Laurie, V.F.; Ricci, A.; Laghi, L.; Parpinello, G.P. Progress in Authentication, Typification and Traceability of Grapes and Wines by Chemometric Approaches; Elsevier Inc.: Amsterdam, The Netherlands, 2014; Volume 60, ISBN 3904573381. [Google Scholar]
- Morata, A. Red Wine Technology; Academic Press: Cambridge, MA, USA, 2018; ISBN 9780128144008. [Google Scholar]
- Alañón, M.E.; Pérez-Coello, M.S.; Marina, M.L. Wine science in the metabolomics era. TrAC Trends Anal. Chem. 2015, 74, 1–20. [Google Scholar] [CrossRef]
- Cozzolino, D. Metabolomics in grape and wine: Definition, current status and future prospects. Food Anal. Methods 2016, 9, 2986–2997. [Google Scholar] [CrossRef]
- Zeng, L.; Pons-Mercadé, P.; Richard, T.; Krisa, S.; Teissèdre, P.-L.; Jourdes, M. Crown procyanidin tetramer: A procyanidin with an unusual cyclic skeleton with a potent protective effect against amyloid-β-induced toxicity. Molecules 2019, 24, 1915. [Google Scholar] [CrossRef] [Green Version]
- Arapitsas, P.; Ugliano, M.; Perenzoni, D.; Angeli, A.; Pangrazzi, P.; Mattivi, F. Wine metabolomics reveals new sulfonated products in bottled white wines, promoted by small amounts of oxygen. J. Chromatogr. A 2016, 1429, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Roullier-Gall, C.; Witting, M.; Gougeon, R.D.; Schmitt-Kopplin, P. High precision mass measurements for wine metabolomics. Front. Chem. 2014, 2, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef]
- Arbulu, M.; Sampedro, M.C.; Gómez-Caballero, A.; Goicolea, M.A.; Barrio, R.J. Untargeted metabolomic analysis using liquid chromatography quadrupole time-of-flight mass spectrometry for non-volatile profiling of wines. Anal. Chim. Acta 2015, 858, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Schueuermann, C.; Steel, C.C.; Blackman, J.W.; Clark, A.C.; Schwarz, L.J.; Moraga, J.; Collado, I.G.; Schmidtke, L.M. A GC–MS untargeted metabolomics approach for the classification of chemical differences in grape juices based on fungal pathogen. Food Chem. 2019, 270, 375–384. [Google Scholar] [CrossRef]
- Gil, M.; Reynes, C.; Cazals, G.; Enjalbal, C.; Sabatier, R.; Saucier, C. Discrimination of rosé wines using shotgun metabolomics with a genetic algorithm and MS ion intensity ratios. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arapitsas, P.; Speri, G.; Angeli, A.; Perenzoni, D.; Mattivi, F. The influence of storage on the “chemical age” of red wines. Metabolomics 2014, 10, 816–832. [Google Scholar] [CrossRef]
- Roullier-Gall, C.; Kanawati, B.; Hemmler, D.; Druschel, G.K.; Gougeon, R.D.; Schmitt-Kopplin, P. Electrochemical triggering of the Chardonnay wine metabolome. Food Chem. 2019, 286, 64–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-K.; Lan, Y.-B.; Huang, Y.; Zhao, X.; Duan, C.-Q. Targeted metabolomics of anthocyanin derivatives during prolonged wine aging: Evolution, color contribution and aging prediction. Food Chem. 2021, 339, 127795. [Google Scholar] [CrossRef]
- Ontañón, I.; Sánchez, D.; Sáez, V.; Mattivi, F.; Ferreira, V.; Arapitsas, P. Liquid chromatography-mass spectrometry-based metabolomics for understanding the compositional changes induced by oxidative or anoxic storage of red wines. J. Agric. Food Chem. 2020, 68, 13367–13379. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; De Rosso, M.; Dalla Vedova, A.; Flamini, R. High-Resolution mass spectrometry identification of secondary metabolites in four red grape varieties potentially useful as traceability markers of wines. Beverages 2018, 4, 74. [Google Scholar] [CrossRef] [Green Version]
- Millán, L.; Sampedro, M.C.; Sánchez, A.; Delporte, C.; Van Antwerpen, P.; Goicolea, M.A.; Barrio, R.J. Liquid chromatography–quadrupole time of flight tandem mass spectrometry–based targeted metabolomic study for varietal discrimination of grapes according to plant sterols content. J. Chromatogr. A 2016, 1454, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Rocchetti, G.; Gatti, M.; Bavaresco, L.; Lucini, L. Untargeted metabolomics to investigate the phenolic composition of Chardonnay wines from different origins. J. Food Compos. Anal. 2018, 71, 87–93. [Google Scholar] [CrossRef]
- Barnaba, C.; Dellacassa, E.; Nicolini, G.; Nardin, T.; Serra, M.; Larcher, R. Non-targeted glycosidic profiling of international wines using neutral loss-high resolution mass spectrometry. J. Chromatogr. A 2018, 1557, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R. Grape and wine metabolomics to develop new insights using untargeted and targeted approaches. Fermentation 2018, 4, 92. [Google Scholar] [CrossRef] [Green Version]
- Lacalle-Bergeron, L.; Izquierdo-Sandoval, D.; Sancho, J.V.; López, F.J.; Hernández, F.; Portolés, T. Chromatography hyphenated to high resolution mass spectrometry in untargeted metabolomics for investigation of food (BIO) markers. TrAC Trends Anal. Chem. 2020, 135, 116161. [Google Scholar] [CrossRef]
- Vrhovsek, U.; Masuero, D.; Gasperotti, M.; Franceschi, P.; Caputi, L.; Viola, R.; Mattivi, F. A versatile targeted metabolomics method for the rapid quantification of multiple classes of phenolics in fruits and beverages. J. Agric. Food Chem. 2012, 60, 8831–8840. [Google Scholar] [CrossRef]
- Barnaba, C.; Dellacassa, E.; Nicolini, G.; Nardin, T.; Malacarne, M.; Larcher, R. Identification and quantification of 56 targeted phenols in wines, spirits, and vinegars by online solid-phase extraction-ultrahigh-performance liquid chromatography-quadrupole-orbitrap mass spectrometry. J. Chromatogr. A 2015, 1423, 124–135. [Google Scholar] [CrossRef]
- Flamini, R.; De Rosso, M.; Bavaresco, L. Study of grape polyphenols by liquid chromatography-high-resolution mass spectrometry (UHPLC/QTOF) and suspect screening analysis. J. Anal. Methods Chem. 2015. [Google Scholar] [CrossRef]
- De Rosso, M.; Mayr, C.M.; Girardi, G.; Vedova, A.D.; Flamini, R. High-resolution mass spectrometry metabolomics of grape chemical markers to reveal use of not-allowed varieties in the production of Amarone and Recioto wines. Metabolomics 2018, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kallithraka, S.; Tsoutsouras, E.; Tzourou, E.; Lanaridis, P. Principal phenolic compounds in Greek red wines. Food Chem. 2006, 99, 784–793. [Google Scholar] [CrossRef]
- Sakkiadi, A.V.; Stavrakakis, M.N.; Haroutounian, S.A. Direct HPLC assay of five biologically interesting phenolic antioxidants in varietal Greek red wines. LWT Food Sci. Technol. 2001, 34, 410–413. [Google Scholar] [CrossRef]
- Karimali, D.; Kosma, I.; Badeka, A. Varietal classification of red wine samples from four native Greek grape varieties based on volatile compound analysis, color parameters and phenolic composition. Eur. Food Res. Technol. 2020, 246, 41–53. [Google Scholar] [CrossRef]
- Tzachristas, A.; Pasvanka, K.; Calokerinos, A.; Proestos, C. Polyphenols: Natural antioxidants to be used as a quality tool in wine authenticity. Appl. Sci. 2020, 10, 5908. [Google Scholar] [CrossRef]
- Jackson, R.S. Wine Science: Principles and Applications; Elsevier Inc.: Amsterdam, The Netherlands, 2020; ISBN 0128165375. [Google Scholar]
- Piñeiro, Z.; Cantos-Villar, E.; Palma, M.; Puertas, B. Direct liquid chromatography method for the simultaneous quantification of hydroxytyrosol and tyrosol in red wines. J. Agric. Food Chem. 2011, 59, 11683–11689. [Google Scholar] [CrossRef] [PubMed]
- Flamini, R.; Mattivi, F.; De Rosso, M.; Arapitsas, P.; Bavaresco, L. Advanced knowledge of three important classes of grape phenolics: Anthocyanins, stilbenes and flavonols. Int. J. Mol. Sci. 2013, 14, 19651–19669. [Google Scholar] [CrossRef]
- Arapitsas, P.; Guella, G.; Mattivi, F. The impact of SO2 on wine flavanols and indoles in relation to wine style and age. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gawel, R.; Day, M.; Van Sluyter, S.C.; Holt, H.; Waters, E.J.; Smith, P.A. White wine taste and mouthfeel as affected by juice extraction and processing. J. Agric. Food Chem. 2014, 62, 10008–10014. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liang, F.; Bin, Y.; Li, P.; Duan, C. Screening non-colored phenolics in red wines using liquid chromatography/ultraviolet and mass spectrometry/mass spectrometry libraries. Molecules 2007, 12, 679–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyraleou, M.; Kallithraka, S.; Gkanidi, E.; Koundouras, S.; Mannion, D.T.; Kilcawley, K.N. Discrimination of five Greek red grape varieties according to the anthocyanin and proanthocyanidin profiles of their skins and seeds. J. Food Compos. Anal. 2020, 92, 103547. [Google Scholar] [CrossRef]
- Kallithraka, S.; Arvanitoyannis, I.; Kefalas, P.; El-Zajouli, A.; Soufleros, E.; Psarra, E. Instrumental and sensory analysis of Greek wines; implementation of principal component analysis (PCA) for classification according to geographical origin. Food Chem. 2001, 73, 501–514. [Google Scholar] [CrossRef]
- Sun, J.; Lin, L.Z.; Chen, P. Study of the mass spectrometric behaviors of anthocyanins in negative ionization mode and its applications for characterization of anthocyanins and non-anthocyanin polyphenols. Rapid Commun. Mass Spectrom. 2012, 26, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Bouloumpasi, E.; Soufleros, E.H.; Tsarchopoulos, C.; Biliaderis, C.G. Primary amino acid composition and its use in discrimination of greek red wines with regard to variety and cultivation region. Vitis 2002, 41, 195–202. [Google Scholar]
- Lambert, M.; Meudec, E.; Verbaere, A.; Mazerolles, G.; Wirth, J.; Masson, G.; Cheynier, V.; Sommerer, N. A high-throughput UHPLC-QqQ-MS method for polyphenol profiling in rosé wines. Molecules 2015, 20, 7890–7914. [Google Scholar] [CrossRef] [PubMed]
- Dasenaki, M.E.; Drakopoulou, S.K.; Aalizadeh, R.; Thomaidis, N.S. Targeted and untargeted metabolomics as an enhanced tool for the detection of pomegranate juice adulteration. Foods 2019, 8, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Baltenweck-Guyot, R.; Trendel, J.M.; Albrecht, P.; Schaeffer, A. Glycosides and phenylpropanoid glycerol in Vitis vinifera Cv. Gewurztraminer wine. J. Agric. Food Chem. 2000, 48, 6178–6182. [Google Scholar] [CrossRef] [PubMed]
- Aalizadeh, R.; Thomaidis, N.S.; Bletsou, A.A.; Gago-Ferrero, P. Quantitative structure-retention relationship models to support nontarget high-resolution mass spectrometric screening of emerging contaminants in environmental samples. J. Chem. Inf. Model. 2016, 56, 1384–1398. [Google Scholar] [CrossRef]
- Kalogiouri, N.P.; Aalizadeh, R.; Thomaidis, N.S. Investigating the organic and conventional production type of olive oil with target and suspect screening by LC-QTOF-MS, a novel semi-quantification method using chemical similarity and advanced chemometrics. Anal. Bioanal. Chem. 2017, 409, 5413–5426. [Google Scholar] [CrossRef]
- Kalogiouri, N.P.; Alygizakis, N.A.; Aalizadeh, R.; Thomaidis, N.S. Olive oil authenticity studies by target and nontarget LC–QTOF-MS combined with advanced chemometric techniques. Anal. Bioanal. Chem. 2016, 408, 7955–7970. [Google Scholar] [CrossRef]
- Gago-Ferrero, P.; Schymanski, E.L.; Bletsou, A.A.; Aalizadeh, R.; Hollender, J.; Thomaidis, N.S. Extended suspect and non-target strategies to characterize emerging polar organic contaminants in raw wastewater with LC-HRMS/MS. Environ. Sci. Technol. 2015, 49, 12333–12341. [Google Scholar] [CrossRef] [Green Version]
- Aalizadeh, R.; Nika, M.C.; Thomaidis, N.S. Development and application of retention time prediction models in the suspect and non-target screening of emerging contaminants. J. Hazard. Mater. 2019, 363, 277–285. [Google Scholar] [CrossRef]
- Djoumbou-Feunang, Y.; Pon, A.; Karu, N.; Zheng, J.; Li, C.; Arndt, D.; Gautam, M.; Allen, F.; Wishart, D.S. CFM-ID 3.0: Significantly Improved ESI-MS/MS Prediction and Compound Identification. Metabolites 2019, 9, 72. [Google Scholar] [CrossRef] [Green Version]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Kalogiouri, N.P.; Aalizadeh, R.; Dasenaki, M.E.; Thomaidis, N.S. Authentication of Greek PDO kalamata table olives: A novel non-target high resolution mass spectrometric approach. Molecules 2020, 25, 2919. [Google Scholar] [CrossRef] [PubMed]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Mehmood, T.; Liland, K.H.; Snipen, L.; Sæbø, S. A review of variable selection methods in partial least squares regression. Chemom. Intell. Lab. Syst. 2012, 118, 62–69. [Google Scholar] [CrossRef]






| Group | Agiorgitiko (n = 27) | Xinomavro (n = 19) | All Samples (n = 46) | |||
|---|---|---|---|---|---|---|
| Concentration (mg/L) | ||||||
| mean | range | Mean | range | mean | range | |
| 4-Hydroxybenzoic acid | 0.52 a | 0.14–1.8 | 0.34 a | 0.13–0.71 | 0.44 | 0.13–1.8 |
| Caffeic acid | 2.4 a | 0.98–5.4 | 2.4 a | 0.79–5.1 | 2.4 | 0.79–5.4 |
| Catechin | 27 a | 18–38 | 65 b | 44–83 | 43 | 18–83 |
| Epicatechin | 15 a | 8.0–26 | 38 b | 18–59 | 24 | 8.0–59 |
| Eriodictyol | traces | traces | traces | |||
| Ferulic acid | 0.22 a | <LOD–0.66 | 0.34 a | 0.24–0.47 | 0.27 | <LOD–0.66 |
| Gallic acid | 26.4 a | 11.1–46 | 43 b | 27–64 | 33 | 11–64 |
| Gentisic acid | 0.481 a | 0.12–0.92 | 0.43 a | 0.11–0.71 | 0.46 | 0.11–0.90 |
| Hydroxytyrosol | 0.992 a | 0.72–1.4 | 2.7 b | 1.7–3.9 | 1.7 | 0.72–3.9 |
| Luteolin | traces | traces | traces | |||
| Myricetin | 2.4 a | 0.86–4.5 | 4.5 b | 1.1–10 | 3.3 | 0.86–10 |
| Naringenin | 0.10 a | <LOD–0.16 | 0.072 b | <LOD–0.13 | 0.091 | <LOD–0.16 |
| p-Coumaric acid | 1.80 a | 0.65–5.45 | 1.02 b | <LOD–4.0 | 1.48 | <LOD–5. 5 |
| Protocatechuic acid | 3.0 a | 1.0–7.7 | 1.3 b | 0.28–2.4 | 2.4 | 0.28–7.7 |
| Quercetin | 2.7 a | 1.1–5.3 | 6.2 b | 0.29–10 | 4.2 | 0.29–10 |
| Resveratrol | 1.4 a | 0.21–2.6 | 0.78 a | 0.41–1.7 | 1.1 | 0.21–2.6 |
| Salicylic acid | 0.67 a | 0.13–1.5 | 0.26 b | 0.17–0.51 | 0.50 | 0.13–1.5 |
| Syringic acid | 4.7 a | 0.98–7.4 | 2.1 b | 1.2–3.7 | 3.6 | 0.98–7.4 |
| Taxifolin | 0.10 a | <LOQ–0.17 | 0.11 a | 0.060–0.20 | 0.11 | <LOQ–0.20 |
| Tyrosol | 28 a | 13–43 | 43 b | 17–74 | 34 | 12–74 |
| Vanillic acid | 1.9 a | 0.81–3.8 | 0.99 b | 0.6–1.8 | 1.5 | 0.60–3.8 |
| Vanillin | traces | traces | traces | |||
| Molecular Formula | Proposed Compound | Class | p1 | pcorr1 | VIP | |
|---|---|---|---|---|---|---|
| 1 | C15H14O6 | Catechin | Flavan-3-ols | 20.44 | 0.51 | 3.67 |
| 2 | C9H10O5 | Ethyl gallate | Benzoates | 16.17 | 0.31 | 2.90 |
| 3 | C15H14O6 | Epicatechin | Flavan-3-ols | 15.98 | 0.54 | 2.87 |
| 4 | C17H21NO8 | Indolelactic acid glycoside | Amines | 15.53 | 0.79 | 2.79 |
| 5 | C11H12O3 | Ethyl coumarate | Cinnamates | −15.30 | −0.64 | 2.75 |
| 6 | C5H9NO2 | L-Proline | Amino acids | 14.17 | 0.32 | 2.54 |
| 7 | C23H26O13 | Quercetin 3,3′-dimethyl ether 4′-glucoside | Flavonols | −11.79 | −0.70 | 2.12 |
| 8 | C13H12O8 | Coutaric acid | Cinnamates | −9.50 | −0.53 | 1.71 |
| 9 | C6H13NO2 | (Iso)leucine | Amino acids | −8.02 | −0.45 | 1.44 |
| 10 | C7H6O3 | Salicylic acid | Benzoates | −7.75 | −0.50 | 1.39 |
| 11 | C30H26O12 | Procyanidin dimer 1 | Flavan-3-ols | 7.67 | 0.35 | 1.38 |
| 12 | C21H18O13 | Quercetin 3-glucuronide | Flavonols | −7.33 | −0.48 | 1.32 |
| 13 | C23H24O12 | Malvidin 3-0 Glucoside [M-2H] | Anthocyanins | −7.30 | −0.66 | 1.31 |
| 14 | C18H26O10 | Benzyl O-[arabinofuranosyl-(1->6)-glucoside] | Benzyl alcohol derivatives | −7.19 | −0.44 | 1.29 |
| 15 | C9H10O4 | Protocatechuic acid ethyl ester | Benzoates | −7.13 | −0.45 | 1.28 |
| 16 | C15H14O7 | Gallocatechin | Flavan-3-ols | 6.90 | 0.48 | 1.24 |
| 17 | C7H6O2 | p-Hydroxybenzaldehyde | Benzaldehydes | 5.78 | 0.55 | 1.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tzachristas, A.; Dasenaki, M.E.; Aalizadeh, R.; Thomaidis, N.S.; Proestos, C. Development of a Wine Metabolomics Approach for the Authenticity Assessment of Selected Greek Red Wines. Molecules 2021, 26, 2837. https://doi.org/10.3390/molecules26102837
Tzachristas A, Dasenaki ME, Aalizadeh R, Thomaidis NS, Proestos C. Development of a Wine Metabolomics Approach for the Authenticity Assessment of Selected Greek Red Wines. Molecules. 2021; 26(10):2837. https://doi.org/10.3390/molecules26102837
Chicago/Turabian StyleTzachristas, Alexandros, Marilena E. Dasenaki, Reza Aalizadeh, Nikolaos S. Thomaidis, and Charalampos Proestos. 2021. "Development of a Wine Metabolomics Approach for the Authenticity Assessment of Selected Greek Red Wines" Molecules 26, no. 10: 2837. https://doi.org/10.3390/molecules26102837
APA StyleTzachristas, A., Dasenaki, M. E., Aalizadeh, R., Thomaidis, N. S., & Proestos, C. (2021). Development of a Wine Metabolomics Approach for the Authenticity Assessment of Selected Greek Red Wines. Molecules, 26(10), 2837. https://doi.org/10.3390/molecules26102837