Assessing the Inhibitory Potential of Kinase Inhibitors In Vitro: Major Pitfalls and Suggestions for Improving Comparability of Data Using CK1 Inhibitors as an Example
Abstract
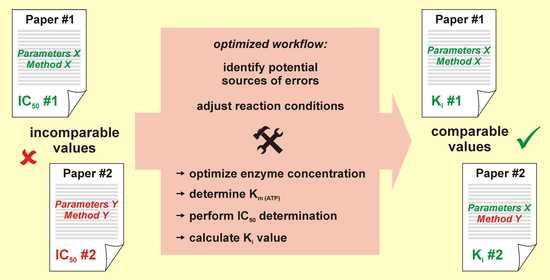
:1. Introduction
1.1. Determination of the Initial Velocity Region
1.2. Determination of Km (ATP) and Ki under Initial Velocity Conditions
2. Results
2.1. Determination of Initial Velocity Region for GST-Tagged CK1δ
2.2. Determination of Initial Velocity Region for 6×His-Tagged CK1δ
2.3. Determination of Km under Initial Velocity Conditions
2.4. Determination of IC50 Values Using Adapted ATP Concentrations
2.5. Determination and Comparison of Kic for GST-CK1δ and 6×His-CK1ε Using the CK1ε Specific Inhibitor PF-4800567
3. Discussion
4. Material and Methods
4.1. Expression and Purification of GST-Tagged CK1δ
4.2. Expression and Purification of 6×His-Tagged CK1δ and 6×His-Tagged CK1ε
4.3. In Vitro Kinase Reactions
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varedi, K.S.M.; Ventura, A.C.; Merajver, S.D.; Lin, X.N. Multisite phosphorylation provides an effective and flexible mechanism for switch-like protein degradation. PLoS ONE 2010, 5, e14029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.-C.; Qi, R.Z.; Paudel, H.; Zhu, H.-J. Regulation and function of protein kinases and phosphatases. Enzyme Res. 2011, 2011, 794089. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, R.; Hanson-Painton, O.; Cooper, D.R. Protein kinases as therapeutic targets. Pharm. Res. 2000, 17, 1345–1353. [Google Scholar] [CrossRef]
- Benn, C.L.; Dawson, L.A. Clinically Precedented Protein Kinases: Rationale for Their Use in Neurodegenerative Disease. Front. Aging Neurosci. 2020, 12, 242. [Google Scholar] [CrossRef]
- Fabbro, D.; Cowan-Jacob, S.W.; Moebitz, H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015, 172, 2675–2700. [Google Scholar] [CrossRef] [Green Version]
- Miljković, F.; Bajorath, J. Computational Analysis of Kinase Inhibitors Identifies Promiscuity Cliffs across the Human Kinome. ACS Omega 2018, 3, 17295–17308. [Google Scholar] [CrossRef]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021. [Google Scholar] [CrossRef]
- Mashhoon, N.; DeMaggio, A.J.; Tereshko, V.; Bergmeier, S.C.; Egli, M.; Hoekstra, M.F.; Kuret, J. Crystal structure of a conformation-selective casein kinase-1 inhibitor. J. Biol. Chem. 2000, 275, 20052–20060. [Google Scholar] [CrossRef] [Green Version]
- Rena, G.; Bain, J.; Elliott, M.; Cohen, P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 2004, 5, 60–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bain, J.; Plater, L.; Elliott, M.; Shpiro, N.; Hastie, C.J.; McLauchlan, H.; Klevernic, I.; Arthur, J.S.C.; Alessi, D.R.; Cohen, P. The selectivity of protein kinase inhibitors: A further update. Biochem. J. 2007, 408, 297–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badura, L.; Swanson, T.; Adamowicz, W.; Adams, J.; Cianfrogna, J.; Fisher, K.; Holland, J.; Kleiman, R.; Nelson, F.; Reynolds, L.; et al. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther. 2007, 322, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.M.; Fisher, K.; Rubitski, D.; Marconi, M.; Meng, Q.-J.; Sládek, M.; Adams, J.; Bass, M.; Chandrasekaran, R.; Butler, T.; et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J. Pharmacol. Exp. Ther. 2009, 330, 430–439. [Google Scholar] [CrossRef]
- Myers, S.M.; Bawn, R.H.; Bisset, L.C.; Blackburn, T.J.; Cottyn, B.; Molyneux, L.; Wong, A.-C.; Cano, C.; Clegg, W.; Harrington, R.W.; et al. High-Throughput Screening and Hit Validation of Extracellular-Related Kinase 5 (ERK5) Inhibitors. ACS Comb. Sci. 2016, 18, 444–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashem, M.A.; Nelson, R.M.; Yingling, J.D.; Pullen, S.S.; Prokopowicz, A.S.; Jones, J.W.; Wolak, J.P.; Rogers, G.R.; Morelock, M.M.; Snow, R.J.; et al. Three mechanistically distinct kinase assays compared: Measurement of intrinsic ATPase activity identified the most comprehensive set of ITK inhibitors. J. Biomol. Screen. 2007, 12, 70–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koresawa, M.; Okabe, T. High-throughput screening with quantitation of ATP consumption: A universal non-radioisotope, homogeneous assay for protein kinase. Assay Drug Dev. Technol. 2004, 2, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.R.; Garcia-Barrio, M.T.; Zhang, X.; Wang, Q.; Taylor, D.R.; Zhang, F.; Herring, C.; Mathews, M.B.; Qin, J.; Hinnebusch, A.G. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol. Cell. Biol. 1998, 18, 2282–2297. [Google Scholar] [CrossRef] [Green Version]
- Cullati, S.; Chen, J.-S.; Gould, K. Autophosphorylation of the CK1 kinase domain regulates enzyme activity and function. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Beenstock, J.; Mooshayef, N.; Engelberg, D. How Do Protein Kinases Take a Selfie (Autophosphorylate)? Trends Biochem. Sci. 2016, 41, 938–953. [Google Scholar] [CrossRef] [PubMed]
- Bhoir, S.; Shaik, A.; Thiruvenkatam, V.; Kirubakaran, S. High yield bacterial expression, purification and characterisation of bioactive Human Tousled-like Kinase 1B involved in cancer. Sci. Rep. 2018, 8, 4796. [Google Scholar] [CrossRef]
- Akizuki, K.; Toyama, T.; Yamashita, M.; Sugiyama, Y.; Ishida, A.; Kameshita, I.; Sueyoshi, N. Facile preparation of highly active casein kinase 1 using Escherichia coli constitutively expressing lambda phosphatase. Anal. Biochem. 2018, 549, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Gietzen, K.F.; Virshup, D.M. Identification of inhibitory autophosphorylation sites in casein kinase I epsilon. J. Biol. Chem. 1999, 274, 32063–32070. [Google Scholar] [CrossRef] [Green Version]
- Budini, M.; Jacob, G.; Jedlicki, A.; Pérez, C.; Allende, C.C.; Allende, J.E. Autophosphorylation of carboxy-terminal residues inhibits the activity of protein kinase CK1alpha. J. Cell. Biochem. 2009, 106, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Brooks, H.B.; Geeganage, S.; Kahl, S.D.; Montrose, C.; Sittampalam, S.; Smith, M.C.; Weidner, J.R. Basics of Enzymatic Assays for HTS. In Assay Guidance Manual; Markossian, S., Sittampalam, G.S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Caaveiro, J.M., Chung, T.D., et al., Eds.; Eli Lilly & Company: Indianapolis, IN, USA; The National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Wang, Y.; Guan, J.; Di Trani, J.M.; Auclair, K.; Mittermaier, A.K. Inhibition and Activation of Kinases by Reaction Products: A Reporter-Free Assay. Anal. Chem. 2019, 91, 11803–11811. [Google Scholar] [CrossRef] [PubMed]
- Knight, Z.A.; Shokat, K.M. Features of selective kinase inhibitors. Chem. Biol. 2005, 12, 621–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knippschild, U.; Gocht, A.; Wolff, S.; Huber, N.; Löhler, J.; Stöter, M. The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell. Signal. 2005, 17, 675–689. [Google Scholar] [CrossRef]
- Xu, P.; Ianes, C.; Gärtner, F.; Liu, C.; Burster, T.; Bakulev, V.; Rachidi, N.; Knippschild, U.; Bischof, J. Structure, regulation, and (patho-)physiological functions of the stress-induced protein kinase CK1 delta (CSNK1D). Gene 2019, 715, 144005. [Google Scholar] [CrossRef] [PubMed]
- Knippschild, U.; Krüger, M.; Richter, J.; Xu, P.; García-Reyes, B.; Peifer, C.; Halekotte, J.; Bakulev, V.; Bischof, J. The CK1 Family: Contribution to Cellular Stress Response and Its Role in Carcinogenesis. Front. Oncol. 2014, 4, 96. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Witt, L.; Ianes, C.; Bischof, J.; Bammert, M.-T.; Baier, J.; Kirschner, S.; Henne-Bruns, D.; Xu, P.; Kornmann, M.; et al. Newly Developed CK1-Specific Inhibitors Show Specifically Stronger Effects on CK1 Mutants and Colon Cancer Cell Lines. Int. J. Mol. Sci. 2019, 20, 184. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A. The mechanism of enzyme-inhibitor-substrate reactions: Illustrated by the cholinesterase-physostigmine-acetylcholine system. J. Gen. Physiol. 1944, 27, 529–580. [Google Scholar] [CrossRef] [Green Version]
- Wolf, A.; Shimamura, S.; Reinhard, F.B.M. Working with small molecules: Preparing and storing stock solutions and determination of kinetic solubility. Methods Mol. Biol. 2012, 803, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Komori, T.; Ishibashi, K.; Uto, T.; Kobayashi, I.; Kadokawa, R.; Kato, Y.; Ninomiya, K.; Takahashi, K.; Hirata, E. Non-aqueous, zwitterionic solvent as an alternative for dimethyl sulfoxide in the life sciences. Commun. Chem. 2020, 3. [Google Scholar] [CrossRef]
- Rivers, A.; Gietzen, K.F.; Vielhaber, E.; Virshup, D.M. Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J. Biol. Chem. 1998, 273, 15980–15984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graves, P.R.; Roach, P.J. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 1995, 270, 21689–21694. [Google Scholar] [CrossRef] [Green Version]
- Bischof, J.; Leban, J.; Zaja, M.; Grothey, A.; Radunsky, B.; Othersen, O.; Strobl, S.; Vitt, D.; Knippschild, U. 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1δ/ε. Amino Acids 2012, 43, 1577–1591. [Google Scholar] [CrossRef] [Green Version]
- Richter, J.; Bischof, J.; Zaja, M.; Kohlhof, H.; Othersen, O.; Vitt, D.; Alscher, V.; Pospiech, I.; García-Reyes, B.; Berg, S.; et al. Difluoro-dioxolo-benzoimidazol-benzamides As Potent Inhibitors of CK1δ and ε with Nanomolar Inhibitory Activity on Cancer Cell Proliferation. J. Med. Chem. 2014, 57, 7933–7946. [Google Scholar] [CrossRef] [PubMed]
- Kusuda, J.; Hidari, N.; Hirai, M.; Hashimoto, K. Sequence analysis of the cDNA for the human casein kinase I delta (CSNK1D) gene and its chromosomal localization. Genomics 1996, 32, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Fish, K.J.; Cegielska, A.; Getman, M.E.; Landes, G.M.; Virshup, D.M. Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J. Biol. Chem. 1995, 270, 14875–14883. [Google Scholar] [CrossRef] [Green Version]










| Working Group A | Working Group B | |||||||
|---|---|---|---|---|---|---|---|---|
| Inhibitor | IC50 [µM] | ATP [µM] | Isoform | Ref. | IC50 [µM] | ATP [µM] | Isoform | Ref. |
| IC261 | 1 | 10 | CK1δ | [9] | 2.5 | 100 | CK1δ ** | [10] |
| D4476 | 0.3 | 20 | CK1δ * | [11] | 0.2 | 100 | CK1δ ** | [10] |
| PF-670462 | 0.008 | 5 | CK1ε * | [12] | 0.08 | 10 | CK1ε | [13] |
| (A) | ||||
| Enzyme | Inhibitor | IC50 [nM] | Kic [nM] | IC50 in Literature [nM] |
| GST-CK1δ | PF-670462 | 69.85 | 34.93 | 13 * [13] |
| Liu-20 | 395.80 | 197.90 | 86 [30] | |
| PF-4800567 | 2012.00 | 1006.00 | 711 * [13] | |
| 6×His-CK1δ | PF-670462 | 64.18 | 32.09 | - |
| Liu-20 | 403.60 | 201.80 | - | |
| 6×His-CK1ε | PF-4800567 | 72.30 | 36.15 | 32 ** [13] |
| (B) | ||||
| Inhibitor | PF-670462 | PF-4800567 | Liu-20 | |
| Structure |  |  |  | |
| Reference | [12] | [13] | [30] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roth, A.; Gihring, A.; Göser, F.; Peifer, C.; Knippschild, U.; Bischof, J. Assessing the Inhibitory Potential of Kinase Inhibitors In Vitro: Major Pitfalls and Suggestions for Improving Comparability of Data Using CK1 Inhibitors as an Example. Molecules 2021, 26, 4898. https://doi.org/10.3390/molecules26164898
Roth A, Gihring A, Göser F, Peifer C, Knippschild U, Bischof J. Assessing the Inhibitory Potential of Kinase Inhibitors In Vitro: Major Pitfalls and Suggestions for Improving Comparability of Data Using CK1 Inhibitors as an Example. Molecules. 2021; 26(16):4898. https://doi.org/10.3390/molecules26164898
Chicago/Turabian StyleRoth, Aileen, Adrian Gihring, Florian Göser, Christian Peifer, Uwe Knippschild, and Joachim Bischof. 2021. "Assessing the Inhibitory Potential of Kinase Inhibitors In Vitro: Major Pitfalls and Suggestions for Improving Comparability of Data Using CK1 Inhibitors as an Example" Molecules 26, no. 16: 4898. https://doi.org/10.3390/molecules26164898
APA StyleRoth, A., Gihring, A., Göser, F., Peifer, C., Knippschild, U., & Bischof, J. (2021). Assessing the Inhibitory Potential of Kinase Inhibitors In Vitro: Major Pitfalls and Suggestions for Improving Comparability of Data Using CK1 Inhibitors as an Example. Molecules, 26(16), 4898. https://doi.org/10.3390/molecules26164898