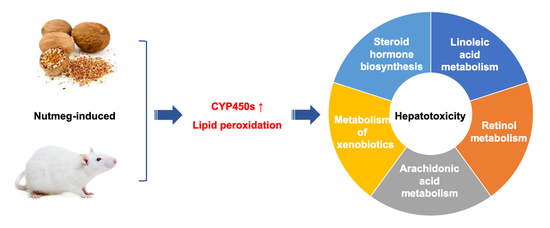
A Proteomics Study on the Mechanism of Nutmeg-Induced Hepatotoxicity
Abstract
:1. Introduction
2. Results
2.1. H&E Staining
2.2. Proteomic Pattern in the Nutmeg Exposure and Control Group
2.2.1. Volcano Plots of Differentially Expressed Proteins (DEPs)
2.2.2. Cluster Analysis of DEPs
2.2.3. DEPs Gene Ontology (GO) Function Enrichment Analysis
2.2.4. Enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
2.2.5. Protein–Protein Interaction (PPI) Network Analysis
2.3. Biochemical Analysis of Serum Monoamine Oxidase (MAO) and Glutathione Peroxidase (GSH-Px), Malondialdehyde (MDA) and Glutathione s-Transferase (GSTs) in Liver Tissue
3. Discussion
4. Materials and Methods
4.1. Chemical Reagent
4.2. Animal Model and Sample Collection
4.3. Hematoxylin and Eosin (H&E) Staining
4.4. Total Protein Extraction
4.5. Protein Quality Test
4.6. TMT Labeling of Peptides
4.7. Separation of Fractions
4.8. LC-MS/MS Analysis
4.9. The Identification and Quantitation of Protein
4.10. The Functional Analysis of Protein and DEP
4.11. Validation of the Biomarkers of Oxidative Stress by ELISA
4.12. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Payne, R.B. Nutmeg Intoxication. N. Engl. J. Med. 1963, 269, 36–38. [Google Scholar] [CrossRef]
- Carstairs, S.D.; Cantrell, F.L. The spice of life: An analysis of nutmeg exposures in California. Clin. Toxicol. 2011, 49, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Beckerman, B.; Persaud, H. Nutmeg overdose: Spice not so nice. Complement. Ther. Med. 2019, 46, 44–46. [Google Scholar] [CrossRef]
- Hayfaa, A.A.-S.; Sahar, A.M.A.-S.; Awatif, M.A.-S. Evaluation of analgesic activity and toxicity of alkaloids in Myristica fragrans seeds in mice. J. Pain Res. 2013, 6, 611–615. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Nordt, S.P.; Ryan, J. Acute nutmeg poisoning. Eur. J. Emerg. Med. 2004, 11, 240–241. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, P.; Guo, X.; Kang, W. Procoagulant substance and mechanism of myristica fragrans. J. Med. Food 2016, 19, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Forrester, M.B. Nutmeg intoxication in Texas, 1998–2004. Hum. Exp. Toxicol. 2005, 24, 563–566. [Google Scholar] [CrossRef]
- Gopalakrishnan, M. Chemical composition of nutmeg and mace. J. Spices Arom. Crops 1992, 1, 49–54. [Google Scholar]
- Gupta, A.D.; Bansal, V.K.; Babu, V.; Maithil, N. Chemistry, antioxidant and antimicrobial potential of nutmeg (Myristica fragrans Houtt). J. Genet. Eng. Biotechnol. 2013, 11, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, P.H.; Le, T.V.T.; Kang, H.W.; Chae, J.; Kim, S.K.; Kwon, K.-I.; Seo, D.B.; Lee, S.J.; Oh, W.K. AMP-activated protein kinase (AMPK) activators from Myristica fragrans (nutmeg) and their anti-obesity effect. Bioorganic Med. Chem. Lett. 2010, 20, 4128–4131. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Jinno, K.; Kawagishi, H.; Arimoto, Y.; Suganuma, H.; Inakuma, T.; Sugiyama, K. Hepatoprotective effect of myristicin from nutmeg (Myristica fragrans) On lipopolysaccharide/D-galactosamine-induced liver injury. J. Agric. Food Chem. 2003, 51, 1560–1565. [Google Scholar] [CrossRef]
- Yang, X.-N.; Liu, X.-M.; Fang, J.-H.; Zhu, X.; Yang, X.-W.; Xiao, X.-R.; Huang, J.-F.; Gonzalez, F.J.; Li, F. PPARα mediates the hepatoprotective effects of nutmeg. J. Proteome Res. 2018, 17, 1887–1897. [Google Scholar] [CrossRef]
- Beyer, J.; Ehlers, D.; Maurer, H.H. Abuse of nutmeg (Myristica fragrans houtt.): Studies on the metabolism and the toxicologic detection of its ingredients elemicin, myristicin, and safrole in rat and human urine using gas chromatography/Mass spectrometry. Ther. Drug Monit. 2006, 28, 568–575. [Google Scholar] [CrossRef]
- Brenner, O.S.F.N.; Knight, E. Chronic nutmeg psychosis. J. R. Soc. Med. 1993, 86, 179–180. [Google Scholar]
- Van Gils, C.; Cox, P.A. Ethnobotany of nutmeg in the Spice Islands. J. Ethnopharmacol. 1994, 42, 117–124. [Google Scholar] [CrossRef]
- Reynoard, J.; Torrents, R.; Domange, B.; Glaizal, M.; De Haro, L.; Simon, N. Nutmeg poisoning: Ten years (2008–2018) of experience from the Marseille Poison Control Center. Presse Méd. 2019, 48, 994–996. [Google Scholar] [CrossRef]
- Ehrenpreis, J.E.; DesLauriers, C.; Lank, P.; Armstrong, P.K.; Leikin, J.B. Nutmeg poisonings: A retrospective review of 10 years experience from the Illinois poison center, 2001–2011. J. Med. Toxicol. 2014, 10, 148–151. [Google Scholar] [CrossRef] [Green Version]
- Sangalli, B.C.; Chiang, W. Toxicology of nutmeg abuse. J. Toxicol. Clin. Toxicol. 2000, 38, 671–678. [Google Scholar] [CrossRef]
- Singh, D.K.; Jaiswal, D.P.; Kumar, D.P.; Singh, D.V.K. Biological effects of myristica fragrans. Ann. Rev. Biomed. Sci. 2009, 11. [Google Scholar] [CrossRef]
- Cao, Z.; Xia, W.; Zhang, X.; Yuan, H.; Guan, D.; Gao, L. Hepatotoxicity of nutmeg: A pilot study based on metabolomics. Biomed. Pharmacother. 2020, 131, 110780. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Paša-Tolić, L. High-throughput proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454. [Google Scholar] [CrossRef] [Green Version]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and their applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.K.; Kim, J.H.; Jung, J.W.; Choi, J.W.; Han, E.S.; Lee, S.H.; Ko, K.H.; Ryu, J.H. Myristicin-induced neurotoxicity in human neuroblastoma SK-N-SH cells. Toxicol. Lett. 2005, 157, 49–56. [Google Scholar] [CrossRef]
- Mansuy, D. The great diversity of reactions catalyzed by cytochromes P450. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 1998, 121, 5–14. [Google Scholar] [CrossRef]
- Lammert, C.; Bjornsson, E.; Niklasson, A.; Chalasani, N. Oral medications with significant hepatic metabolism at higher risk for hepatic adverse events. Hepatology 2010, 51, 615–620. [Google Scholar] [CrossRef]
- Denisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005, 105, 2253–2278. [Google Scholar] [CrossRef]
- Au, J.S.; Navarro, V.J.; Rossi, S. Review article: Drug-induced liver injury-its pathophysiology and evolving diagnostic tools. Aliment. Pharmacol. Ther. 2011, 34, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Boelsterli, U.A.; Lim, P.L. Mitochondrial abnormalities—A link to idiosyncratic drug hepatotoxicity? Toxicol. Appl. Pharmacol. 2007, 220, 92–107. [Google Scholar] [CrossRef]
- Pessayre, D.; Fromenty, B.; Berson, A.; Robin, M.-A.; Lettéron, P.; Moreau, R.; Mansouri, A. Central role of mitochondria in drug-induced liver injury. Drug Metab. Rev. 2012, 44, 34–87. [Google Scholar] [CrossRef]
- Keys, S.A.; Zimmerman, W.F. Antioxidant activity of retinol, glutathione, and taurine in bovine photoreceptor cell membranes. Exp. Eye Res. 1999, 68, 693–702. [Google Scholar] [CrossRef]
- Kaul, S.; Krishnakantha, T. Influence of retinol deficiency and curcumin/turmeric feeding on tissue microsomal membrane lipid peroxidation and fatty acids in rats. Mol. Cell. Biochem. 1997, 175, 43–48. [Google Scholar] [CrossRef]
- Ross, A.C.; Zolfaghari, R. Regulation of hepatic retinol metabolism: Perspectives from studies on vitamin A status. J. Nutr. 2004, 134, 269S–275S. [Google Scholar] [CrossRef] [PubMed]
- Gutteridge, J.M. Lipid peroxidation and antioxidants as biomarkers of tissue damage. Clin Chem. 1995, 41, 1819–1828. [Google Scholar] [CrossRef] [PubMed]
- Mylonas, C.; Kouretas, D. Lipid peroxidation and tissue damage. In Vivo 1999, 13, 295–309. [Google Scholar] [PubMed]
- Moore, K.; Roberts, L.J. Measurement of lipid peroxidation. Free Radic. Res. 1998, 28, 659–671. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Marnett, L.J. Oxy radicals, lipid peroxidation and DNA damage. Toxicology 2002, 181–182. [Google Scholar] [CrossRef]
- Niu, G.; Zhao, S.; Wang, L.; Dong, W.; Liu, L.; He, Y. Structure of the Arabidopsis thaliana NADPH -cytochrome P450 reductase 2 (ATR2) provides insight into its function. FEBS J. 2017, 284, 754–765. [Google Scholar] [CrossRef] [Green Version]
- Yamamura, Y.; Mabuchi, A. Functional characterization of NADPH-cytochrome P450 reductase and cinnamic acid 4-hydroxylase encoding genes from Scoparia dulcis L. Bot. Stud. 2020, 61, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Huang, J.; Chen, L.; Chen, Y.; Li, X. In vivo and in vitro studies on the roles of p38 mitogen-activated protein kinase and NADPH-cytochrome P450 reductase in Alzheimer’s disease. Exp. Ther. Med. 2017, 14, 4755–4760. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A. Glutathione peroxidase. Meth. Enzymol. 1981, 77, 325–333. [Google Scholar] [CrossRef]
- Chatterjee, A.; Gupta, S. The multifaceted role of glutathione S-transferases in cancer. Cancer Lett. 2018, 433, 33–42. [Google Scholar] [CrossRef]
- McGill, M.R.; Sharpe, M.R.; Williams, C.D.; Taha, M.; Curry, S.C.; Jaeschke, H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J. Clin. Investig. 2012, 122, 1574–1583. [Google Scholar] [CrossRef] [Green Version]
- Watkins, P.B. Role of cytochromes P45O in drug metabolism and hepatotoxicity. Semin. Liver Dis. 1990, 10, 235–250. [Google Scholar] [CrossRef]
- Truitt, E.B., Jr.; Duritz, G.; Ebersberger, E.M. Evidence of Monoamine Oxidase Inhibition by Myristicin and Nutmeg. Proc. Soc. Exp. Biol. Med. 1963, 112, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Loflin, J.; Lopez, N.; Whanger, P.D.; Kioussi, C. Selenoprotein W during development and oxidative stress. J. Inorg. Biochem. 2006, 100, 1679–1684. [Google Scholar] [CrossRef]
- Whanger, P. Selenoprotein expression and function—Selenoprotein W. Biochim. Biophys. Acta (BBA) Gen. Subj. 2009, 1790, 1448–1452. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.-D.; Wu, Q.; Zhang, Z.-W.; Li, S.; Wang, X.-L.; Lei, X.-G.; Xu, S.-W. Selenoprotein W serves as an antioxidant in chicken myoblasts. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 3112–3120. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, W.; Cao, Z.; Zhang, X.; Gao, L. A Proteomics Study on the Mechanism of Nutmeg-Induced Hepatotoxicity. Molecules 2021, 26, 1748. https://doi.org/10.3390/molecules26061748
Xia W, Cao Z, Zhang X, Gao L. A Proteomics Study on the Mechanism of Nutmeg-Induced Hepatotoxicity. Molecules. 2021; 26(6):1748. https://doi.org/10.3390/molecules26061748
Chicago/Turabian StyleXia, Wei, Zhipeng Cao, Xiaoyu Zhang, and Lina Gao. 2021. "A Proteomics Study on the Mechanism of Nutmeg-Induced Hepatotoxicity" Molecules 26, no. 6: 1748. https://doi.org/10.3390/molecules26061748
APA StyleXia, W., Cao, Z., Zhang, X., & Gao, L. (2021). A Proteomics Study on the Mechanism of Nutmeg-Induced Hepatotoxicity. Molecules, 26(6), 1748. https://doi.org/10.3390/molecules26061748