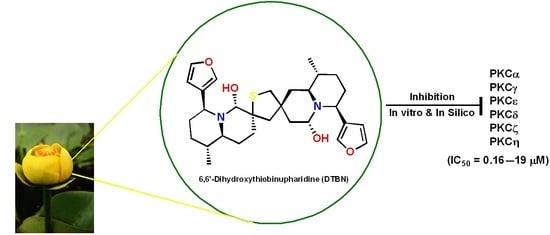
6,6′-Dihydroxythiobinupharidine (DTBN) Purified from Nuphar lutea Leaves Is an Inhibitor of Protein Kinase C Catalytic Activity
Abstract
:1. Introduction
2. Results
2.1. Protein Kinase C (PKC) Inhibition Assay
2.2. Molecular Docking of DTBN to PKC Isoforms
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methodology
4.2.1. In Vitro Kinase Assay
4.2.2. Molecular Docking and MM-GBSA Refinement
4.2.3. Homology Modelling and Structural Validation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Winer, H.; Ozer, J.; Shemer, Y.; Reichenstein, I.; Eilam-Frenkel, B.; Benharroch, D.; Golan-Goldhirsh, A.; Gopas, J. Nuphar lutea Extracts Exhibit Anti-Viral Activity against the Measles Virus. Molecules 2020, 25, 1657. [Google Scholar] [CrossRef] [Green Version]
- Ozer, J.; Levi, T.; Golan-Goldhirsh, A.; Gopas, J. Anti-inflammatory effect of a Nuphar lutea partially purified leaf extract in murine models of septic shock. J. Ethnopharmacol. 2015, 161, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Padgett, D.J. A monograph of nuphar (nymphaeaceae). Rhodora 2007, 109, 1–95. [Google Scholar] [CrossRef]
- Ozer, J.; Eisner, N.; Ostrozhenkova, E.; Bacher, A.; Eisenreich, W.; Benharroch, D.; Golan-Goldhirsh, A.; Gopas, J. Nuphar lutea thioalkaloids inhibit the nuclear factor kappaB pathway, potentiate apoptosis and are synergistic with cisplatin and etoposide. Cancer Biol. Ther. 2009, 8, 1860–1868. [Google Scholar] [CrossRef] [Green Version]
- Yildirim, A.B.; Karakas, F.P.; Turker, A.U. In vitro antibacterial and antitumor activities of some medicinal plant extracts, growing in Turkey. Asian Pac. J. Trop. Med. 2013, 6, 616–624. [Google Scholar] [CrossRef]
- Okamura, S.; Nishiyama, E.; Yamazaki, T.; Otsuka, N.; Taniguchi, S.; Ogawa, W.; Hatano, T.; Tsuchiya, T.; Kuroda, T. Action mechanism of 6,6′-dihydroxythiobinupharidine from Nuphar japonicum, which showed anti-MRSA and anti-VRE activities. Biochim. Biophys. Acta 2015, 1850, 1245–1252. [Google Scholar] [CrossRef]
- Cullen, W.P.; LaLonde, R.T.; Wang, C.J.; Wong, C.F. Isolation and in vitro antifungal activity of 6,6’-dihydroxythiobinupharidine. J. Pharm. Sci. 1973, 62, 826–827. [Google Scholar] [CrossRef] [PubMed]
- El-On, J.; Ozer, L.; Gopas, J.; Sneir, R.; Golan-Goldhirsh, A. Nuphar lutea: In vitro anti-leishmanial activity against Leishmania major promastigotes and amastigotes. Phytomedicine 2009, 16, 788–792. [Google Scholar] [CrossRef]
- Ozer, L.; El-On, J.; Golan-Goldhirsh, A.; Gopas, J. Leishmania major: Anti-leishmanial activity of Nuphar lutea extract mediated by the activation of transcription factor NF-kappaB. Exp. Parasitol. 2010, 126, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Jacob, M.; Walker, L.; Tekwani, B. Screening North American plant extracts in vitro against Trypanosoma brucei for discovery of new antitrypanosomal drug leads. BMC Complement Altern. Med. 2016, 16, 131. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, H.; Yoshida, K.; Miyagawa, K.; Nemoto, Y.; Asao, Y.; Yoshikawa, M. Nuphar alkaloids with immediately apoptosis-inducing activity from Nuphar pumilum and their structural requirements for the activity. Bioorg. Med. Chem. Lett. 2006, 16, 1567–1573. [Google Scholar] [CrossRef]
- Ozer, J.; Fishman, D.; Eilam, B.; Golan-Goldhirsh, A.; Gopas, J. Anti-Metastatic Effect of Semi-Purified Nuphar Lutea Leaf Extracts. J. Cancer 2017, 8, 1433–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalvie, E.D.; Gopas, J.; Golan-Goldhirsh, A.; Osheroff, N. 6,6′-Dihydroxythiobinupharidine as a poison of human type II topoisomerases. Bioorg. Med. Chem. Lett. 2019, 29, 1881–1885. [Google Scholar] [CrossRef]
- Levy, D.H.; Chapple, I.L.C.; Shapira, L.; Golan-Goldhirsh, A.; Gopas, J.; Polak, D. Nupharidine enhances Aggregatibacter actinomycetemcomitans clearance by priming neutrophils and augmenting their effector functions. J. Clin. Periodontol. 2019, 46, 62–71. [Google Scholar] [CrossRef] [Green Version]
- Tada, N.; Jansen, D.J.; Mower, M.P.; Blewett, M.M.; Umotoy, J.C.; Cravatt, B.F.; Wolan, D.W.; Shenvi, R.A. Synthesis and Sulfur Electrophilicity of the Nuphar Thiaspirane Pharmacophore. ACS Cent. Sci. 2016, 2, 401–408. [Google Scholar] [CrossRef]
- Steinberg, S.F. Structural basis of protein kinase C isoform function. Physiol. Rev. 2008, 88, 1341–1378. [Google Scholar]
- Dowling, C.M.; Kiely, P.A. Targeting Protein Kinase C Downstream of Growth Factor and Adhesion Signalling. Cancers 2015, 7, 1271–1291. [Google Scholar] [CrossRef] [PubMed]
- Antal, C.E.; Hudson, A.M.; Kang, E.; Zanca, C.; Wirth, C.; Stephenson, N.L.; Trotter, E.W.; Gallegos, L.L.; Miller, C.J.; Furnari, F.B.; et al. Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell 2015, 160, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.D.; Chen, S.R.; Chen, H.; Pan, H.L. Bortezomib induces neuropathic pain through protein kinase C-mediated activation of presynaptic NMDA receptors in the spinal cord. Neuropharmacology 2017, 123, 477–487. [Google Scholar] [CrossRef]
- Muraleedharan, A.; Rotem-Dai, N.; Strominger, I.; Anto, N.P.; Isakov, N.; Monsonego, A.; Livneh, E. Protein kinase C eta is activated in reactive astrocytes of an Alzheimer’s disease mouse model: Evidence for its immunoregulatory function in primary astrocytes. Glia 2021, 69, 697–714. [Google Scholar] [CrossRef]
- Alkon, D.L.; Sun, M.K.; Nelson, T.J. PKC signaling deficits: A mechanistic hypothesis for the origins of Alzheimer’s disease. Trends Pharmacol. Sci. 2007, 28, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Livneh, E.; Fishman, D.D. Linking protein kinase C to cell-cycle control. Eur. J. Biochem. 1997, 248, 1–9. [Google Scholar] [CrossRef]
- Newton, A.C. Protein kinase C: Structure, function, and regulation. J. Biol. Chem. 1995, 270, 28495–28498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aley, K.O.; Martin, A.; McMahon, T.; Mok, J.; Levine, J.D.; Messing, R.O. Nociceptor sensitization by extracellular signal-regulated kinases. J. Neurosci. 2001, 21, 6933–6939. [Google Scholar] [CrossRef]
- Perluigi, M.; Barone, E.; Di Domenico, F.; Butterfield, D.A. Aberrant protein phosphorylation in Alzheimer disease brain disturbs pro-survival and cell death pathways. Biochim. Biophys. Acta 2016, 1862, 1871–1882. [Google Scholar] [CrossRef]
- Bizzarri, M.; Dinicola, S.; Bevilacqua, A.; Cucina, A. Broad Spectrum Anticancer Activity of Myo-Inositol and Inositol Hexakisphosphate. Int. J. Endocrinol. 2016, 2016, 5616807. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Kong, C.; Yang, X.; Cui, X.; Lin, X.; Zhang, Z. Protein kinase C-alpha (PKCalpha) modulates cell apoptosis by stimulating nuclear translocation of NF-kappa-B p65 in urothelial cell carcinoma of the bladder. BMC Cancer 2017, 17, 432. [Google Scholar] [CrossRef]
- Leonard, B.; McCann, J.L.; Starrett, G.J.; Kosyakovsky, L.; Luengas, E.M.; Molan, A.M.; Burns, M.B.; McDougle, R.M.; Parker, P.J.; Brown, W.L.; et al. The PKC/NF-kappaB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res. 2015, 75, 4538–4547. [Google Scholar] [CrossRef] [Green Version]
- Staal, J.; Beyaert, R. Inflammation and NF-kappaB Signaling in Prostate Cancer: Mechanisms and Clinical Implications. Cells 2018, 7, 122. [Google Scholar] [CrossRef] [Green Version]
- Cooke, M.; Magimaidas, A.; Casado-Medrano, V.; Kazanietz, M.G. Protein kinase C in cancer: The top five unanswered questions. Mol. Carcinog. 2017, 56, 1531–1542. [Google Scholar] [CrossRef]
- Mut, M.; Amos, S.; Hussaini, I.M. PKC alpha phosphorylates cytosolic NF-kappaB/p65 and PKC delta delays nuclear translocation of NF-kappaB/p65 in U1242 glioblastoma cells. Turk. Neurosurg. 2010, 20, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Wagner, J.; von Matt, P.; Sedrani, R.; Albert, R.; Cooke, N.; Ehrhardt, C.; Geiser, M.; Rummel, G.; Stark, W.; Strauss, A.; et al. Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes. J. Med. Chem. 2009, 52, 6193–6196. [Google Scholar] [CrossRef]
- van Eis, M.J.; Evenou, J.P.; Floersheim, P.; Gaul, C.; Cowan-Jacob, S.W.; Monovich, L.; Rummel, G.; Schuler, W.; Stark, W.; Strauss, A.; et al. 2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes. Bioorg. Med. Chem. Lett. 2011, 21, 7367–7372. [Google Scholar] [CrossRef]
- Bi, K.; Tanaka, Y.; Coudronniere, N.; Sugie, K.; Hong, S.; van Stipdonk, M.J.; Altman, A. Antigen-induced translocation of PKC-theta to membrane rafts is required for T cell activation. Nat. Immunol. 2001, 2, 556–563. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Smalley, T.; Metcalf, R.; Patel, R.; Islam, S.M.A.; Bommareddy, R.R.; Acevedo-Duncan, M. The Atypical Protein Kinase C Small Molecule Inhibitor zeta-Stat, and Its Effects on Invasion through Decreases in PKC-zeta Protein Expression. Front. Oncol. 2020, 10, 209. [Google Scholar] [CrossRef] [Green Version]










| PKC Isoforms | IC50 Values (μg/mL) | IC50 Values (μM) |
|---|---|---|
| PKCα | 0.092 | 0.174 |
| PKCγ | 0.089 | 0.168 |
| PKCε | 7.5 | 14.23 |
| PKCη | >10 | >19 |
| PKCζ | 9.8 | 18.6 |
| PKCδ | >10 | >19 |
| PKC Isoforms | ΔG Bind * (Kcal/mol) of DTBN | Control Molecule | ΔG Bind * (Kcal/mol) |
|---|---|---|---|
| PKCα | −40.83 | NVP-AEB071 | −71.92 |
| PKCγ | −30.74 | GO6983 | −57.18 |
| PKCε | −23.25 | NVP-AEB071 | −44.22 |
| PKCη | −20.73 | 2,6-Naphthyridine | −56.52 |
| PKCζ | −19.61 | - | - |
| PKCδ | −19.0 | NVP-AEB071 | −61.35 |
| PKC Isoforms | H-Bond Interaction | π···π and π···S Interaction |
|---|---|---|
| PKCα | (i) −NH hydrogen of Asn421 and furan ring −O of DTBN (N−H···O, 2.91 Å) (ii) carboxylic oxygen (−OH) of Asp424 and hydrogen of −OH group proximal to thiophene sulphur in DTBN (O−H···O, 2.09 Å) (iii) −CH hydrogen of furan ring in DTBN and carboxylic oxygen (C=O) of Leu345 (C−H···O, 2.43 Å) | π···π interaction between phenyl and furan ring of Tyr419 and DTBN, respectively (2.24 Å) |
| PKCγ | carboxylic oxygen (−OH) of Asp441 and hydrogen of −OH proximal to the thiophene sulfur in DTBN (O−H···O, 2.41 Å) | π···π interaction between phenyl and furan ring of Phe362 and DTBN (4.80 Å). π···S interaction between S and furan ring of Cys636 and DTBN, respectively (4.29 Å). |
| PKCε | (i) side-chain carboxylic oxygen (C=O) of Asp536 and hydrogen of −OH group proximal to thiophene sulphur in DTBN (O−H···O, 1.91 Å) (ii) C−H···O aromatic hydrogen bond between (a) furan ring hydrogen of DTBN and side-chain carboxylic oxygen (C=O) of Asp536 (2.68 Å) and carboxylic oxygen (C=O) of Lys416 (2.79 Å) (b) furan ring hydrogen of DTBN and side-chain carboxylic oxygen (C=O) of amino acid residue Asp699 (2.51 Å) | π···π interaction between phenyl and furan ring of Phe419 and DTBN, respectively (4.90 Å). |
| PKCη | carboxylic oxygen (−OH) of Asp440 and carboxylic oxygen (C=O) of Asp483 involved in hydrogen bond with −OH group proximal to thiophene sulphur in DTBN (two O−H···H, 2.31 and 2.88 Å, respectively) | (i) π···S interaction between phenyl and thiophene sulphur of Phe366 and DTBN, respectively (5.02 Å). (ii) π···S interaction between furan ring and sulphur of DTBN and Met442, respectively (4.72 Å). |
| PKCζ | (i) side-chain hydroxy oxygen of Ser262 and carboxylic oxygen (−OH) of Asp380 involved in hydrogen bond with −OH groups of DTBN (two O−H···O, 1.98 and 1.96 Å, respectively) (ii) phenyl hydrogen of amino acid residue Phe552 and furan oxygen of nupharidine (aromatic C−H···O, 3.23 Å) | - |
| PKCδ | - | - |
| Protein Model | Template | Seq. Identity | Seq. Similarity | Coverage | GQME | Qmean | Range |
|---|---|---|---|---|---|---|---|
| PKCγ | 3IW4.1.A | 76.99% | 0.55 | 0.49 | 0.35 | −1.78 | 345–686 |
| PKCδ | 5f9e.1.A | 72.99% | 0.53 | 0.49 | 0.32 | −0.41 | 343–674 |
| PKCε | 3txo.1.A | 72.02% | 0.53 | 0.46 | 0.32 | −0.32 | 406–733 |
| PKCζ | 5li1.1.A | 84.15% | 0.57 | 0.59 | 0.39 | −0.63 | 246–585 |
| Protein Model | MolProbity Score | Ramachandran Favoured | Ramachandran Outliner | ERRAT Overall Quality Factor | ProSA Z-Score |
|---|---|---|---|---|---|
| PKCγ | 1.94 | 93.53% | 0.59% | 93.91 | −8.0 |
| PKCδ | 1.08 | 96.97% | 0.00% | 89.35 | −8.59 |
| PKCε | 1.31 | 96.32% | 0.61% | 88.56 | −8.06 |
| PKCζ | 0.87 | 97.93% | 0.30% | 92.76 | −9.73 |
| PKC Isoforms | Site Score | Predicted Site Amino Acid Residue Numbers |
|---|---|---|
| PKCγ | 1.042 | 357,358,361,362,365,378,380,399,403,418,434,435,436,437,438,439,440,444,445,448,482,484,485,487,490,497,498,499,631,633,634,635,636,638, 639 |
| PKCε | 1.078 | 414,415,418,419,422,435,437,456,460,470,486,487,488,489,492,493,495,496,499,532,534,536,537,539,549,550,551,552,553,697,698,699,702 |
| PKCζ | 1.044 | 258,259,260,261,262,263,264,266,279,281,314,330,331,332,333,337,339,340,343,376,378,380,381,383,393,394,396,397,414,548,549,551 |
| PKCδ | 1.037 | 355,356,358,359,360,361,362,363,376,378,379,380,381,384,385,388,390,392,393,394,396,397,399,401,411,422,427,428,429,430,434,436,437,440,471,472,473,475,477,478,480,490,491,492,493,494,495,496,497,498,507,633,634,637,644,645,646 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waidha, K.; Anto, N.P.; Jayaram, D.R.; Golan-Goldhirsh, A.; Rajendran, S.; Livneh, E.; Gopas, J. 6,6′-Dihydroxythiobinupharidine (DTBN) Purified from Nuphar lutea Leaves Is an Inhibitor of Protein Kinase C Catalytic Activity. Molecules 2021, 26, 2785. https://doi.org/10.3390/molecules26092785
Waidha K, Anto NP, Jayaram DR, Golan-Goldhirsh A, Rajendran S, Livneh E, Gopas J. 6,6′-Dihydroxythiobinupharidine (DTBN) Purified from Nuphar lutea Leaves Is an Inhibitor of Protein Kinase C Catalytic Activity. Molecules. 2021; 26(9):2785. https://doi.org/10.3390/molecules26092785
Chicago/Turabian StyleWaidha, Kamran, Nikhil Ponnoor Anto, Divya Ram Jayaram, Avi Golan-Goldhirsh, Saravanakumar Rajendran, Etta Livneh, and Jacob Gopas. 2021. "6,6′-Dihydroxythiobinupharidine (DTBN) Purified from Nuphar lutea Leaves Is an Inhibitor of Protein Kinase C Catalytic Activity" Molecules 26, no. 9: 2785. https://doi.org/10.3390/molecules26092785
APA StyleWaidha, K., Anto, N. P., Jayaram, D. R., Golan-Goldhirsh, A., Rajendran, S., Livneh, E., & Gopas, J. (2021). 6,6′-Dihydroxythiobinupharidine (DTBN) Purified from Nuphar lutea Leaves Is an Inhibitor of Protein Kinase C Catalytic Activity. Molecules, 26(9), 2785. https://doi.org/10.3390/molecules26092785