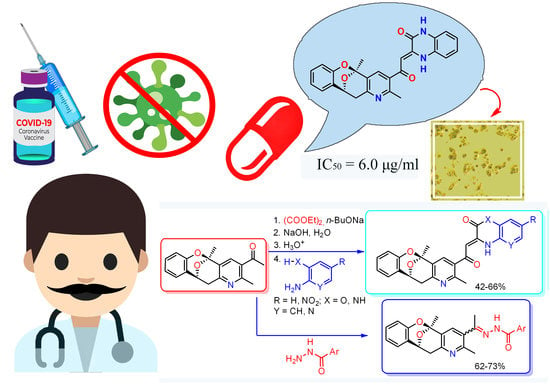
Synthesis and Antiviral Properties against SARS-CoV-2 of Epoxybenzooxocino[4,3-b]Pyridine Derivatives
Abstract
:1. Introduction
2. Results
2.1. Chemistry
2.2. Biological Activity
2.3. Cytotoxicity Studies
2.4. Antiviral Properties
2.5. Methods and Techniques for Molecular Docking
3. Conclusion
4. Materials and Methods
4.1. Synthesis of Compounds
4.2. Biological Tests
4.2.1. Cell Culture and Virus Strain
4.2.2. Virus Stocks
4.2.3. Virus Titers
4.2.4. Control Drug
4.2.5. Cytotoxicity Test
4.2.6. Antiviral Activity
4.3. Data Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Li, J.; Lai, S.; Gao, G.-F.; Shi, W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.; Witek, T.J. The emergency use authorization of pharmaceuticals: History and utility during the COVID-19 pandemic. Pharm. Med. 2021, 35, 203–213. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 updates: Remdesivir (Veklury) in high-risk outpatients with COVID-19. Med. Lett. Drugs Ther. 2022, 64, 31–32.
- COVID-19 updates: NIH outpatient treatment guidelines. Med. Lett. Drugs Ther. 2022, 64, 32.
- Finberg, R.W.; Ashraf, M.; Julg, B.; Ayoade, F.; Marathe, J.G.; Issa, N.C.; Epstein, C. US201 Study: A Phase 2, Randomized Proof-of-Concept Trial of Favipiravir for the Treatment of COVID-19. Open Forum Infect. Dis. 2021, 8, ofab563. [Google Scholar] [CrossRef]
- AlQahtani, M.; Kumar, N.; Aljawder, D.; Abdulrahman, A.; Alnashaba, F.; Fayyad, M.A.; Atkin, S.L. Randomized controlled trial of favipiravir, hydroxychloroquine, and standard care in patients with mild/moderate COVID-19 disease. Sci. Rep. 2022, 12, 1–10. [Google Scholar]
- Kulakov, I.V.; Stalinskaya, A.L.; Chikunov, S.Y.; Gatilov, Y.V. Synthesis of new representatives of 11,12-dihydro-5H-5,11-epoxybenzo[7,8]oxocino[4,3-b] pyridines-structural analogues of integrastatins A, B. New J. Chem. 2021, 45, 3559–3569. [Google Scholar] [CrossRef]
- Oleshchuk, A.L.; Karbainova, A.A.; Krivoruchko, T.N.; Shulgau, Z.T.; Seilkhanov, T.M.; Kulakov, I.V. Synthesis and biological activity of 3, 5-diacetyl-2, 6-dimethylpyridine derivatives. Chem. Heterocycl. Compd. 2019, 55, 47–51. [Google Scholar] [CrossRef]
- Talontsi, F.M.; Dittrich, B.; Schuffler, A.; Sun, H.; Laatsch, H. Epicoccolides: Antimicrobial and antifungal polyketides from an endophytic fungus Epicoccum sp. associated with Theobroma cacao. Eur. J. Org. Chem. 2013, 2013, 3174–3180. [Google Scholar] [CrossRef]
- El Amrani, M.; Lai, D.; Debbab, A.; Aly, A.H.; Siems, K.; Seidel, C.; Schnekenburger, M.; Gaigneaux, A.; Diederich, M.; Feger, D.; et al. Protein kinase and HDAC inhibitors from the endophytic fungus Epicoccum nigrum. J. Nat. Prod. 2014, 77, 49–56. [Google Scholar] [CrossRef]
- Singh, S.B.; Zink, D.L.; Quamina, D.S.; Pelaez, F.; Teran, A.; Felock, P.; Hazuda, D.J. Integrastatins: Structure and HIV-1 integrase inhibitory activities of two novel racemic tetracyclic aromatic heterocycles produced by two fungal species. Tetrahedron Lett. 2002, 43, 2351–2354. [Google Scholar] [CrossRef]
- Oumzil, H.; Ghoulami, S.; Rhajaoui, M.; Ilidrissi, A.; Fkih-Tetouani, S.; Faid, M.; Benjouad, A. Antibacterial and antifungal activity of essential oils of Mentha suaveolens. Phytother. Res. 2002, 16, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, V.; Balaji, S.; Senbagam, R.; Vijayakumar, R.; Rajarajan, M.; Vanangamudi, G.; Thirunarayanan, G. Synthesis and antimicrobial activities of some (E)-N-1-(substituted benzylidene)benzohydrazides. Int. J. Adv. Chem. 2017, 5, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Popiołek, Ł.; Biernasiuk, A. Synthesis and investigation of antimicrobial activities of nitrofurazone analogues containing hydrazide-hydrazone moiety. Saudi Pharm. J. 2017, 25, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Guessas, B.; Othman, A.A.; Khiati, Z. Synthesis and antibacterial activity of 1,3,4-oxadiazole and 1,2,4-triazole derivatives of salicylic acid and its synthetic intermediates. S. Afr. J. Chem. 2007, 60, 20–24. [Google Scholar]
- Vilchèze, C.; Jacobs, W.R., Jr. The mechanism of isoniazid killing: Clarity through the scope of genetics. Annu. Rev. Microbiol. 2007, 61, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Yurttaş, L.; Kaplancıklı, Z.A.; Cantürk, Z.; Gencer, H.K. Synthesis, Antituberculotic, and Cytotoxic Properties of New Hydrazone Derivatives Bearing Pyrimidine-Alkylsulfanyl Moiety. Phosphorus Sulfur Silicon Relat. Elem. 2015, 190, 1183–1191. [Google Scholar] [CrossRef]
- Vinsova, J.; Imramovsky, A.; Jampilek, J.; Monreal, J.; Dolezal, M. Recent advances on isoniazide derivatives. Anti-Infect. Agents Med. Chem. 2008, 7, 12–31. [Google Scholar] [CrossRef]
- Kim, J.; Kang, S.; Hong, S.; Yum, S.; Kim, Y.M.; Jung, Y. Structure–activity relationship of salicylic acid derivatives on inhibition of TNF-α dependent NFκB activity: Implication on anti-inflammatory effect of N-(5-chlorosalicyloyl) phenethylamine against experimental colitis. Eur. J. Med. Chem. 2012, 48, 36–44. [Google Scholar] [CrossRef]
- Pollin, W.; Cardon, P.V.; Kety, S.S. Effects of amino acid feedings in schizophrenic patients treated with iproniazid. Science 1961, 133, 104–105. [Google Scholar] [CrossRef] [Green Version]
- Pi, Y.; Wang, D.-J.; Liu, H.; Hu, Y.-J.; Wei, X.-H.; Zheng, J. Synthesis and spectroscopic properties of some new difluoroboron bis-β-diketonate derivatives. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 131, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Pais, G.C.G.; Zhang, X.; Marchand, C.; Neamati, N.; Cowansage, K.; Svarovskaia, E.S.; Pathak, V.K.; Tang, Y.; Nicklaus, M.; Pommier, Y.; et al. Structure Activity of 3-Aryl-1,3-diketo-Containing Compounds as HIV-1 Integrase Inhibitors. J. Med. Chem. 2002, 45, 3184–3194. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Johnson, A.A.; Marchand, C. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 2005, 4, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Kamath, S.; Sanchez, T.; Neamati, N.; Schinazi, R.F.; Buolamwini, J.K. Synthesis and biological evaluation of novel 5(H)-phenanthridin-6-ones, 5(H)-phenanthridin-6-one diketo acid, and polycyclic aromatic diketo acid analogs as new HIV-1 integrase inhibitors. Bioorg. Med. Chem. 2007, 15, 1212–1228. [Google Scholar] [CrossRef]
- Di Santo, R.; Costi, R.; Roux, A.; Miele, G.; Crucitti, G.C.; Iacovo, A.; Rosi, F.; Lavecchia, A.; Marinelli, L.; di Giovanni, C.; et al. Novel quinolinonyl diketo acid derivatives as HIV-1 integrase inhibitors: Design, synthesis, and biological activities. J. Med. Chem. 2008, 51, 4744–4750. [Google Scholar] [CrossRef] [Green Version]
- Nair, V.; Okello, M. Integrase inhibitor prodrugs: Approaches to enhancing the anti-HIV activity of β-diketo acids. Molecules 2015, 20, 12623–12651. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.P.; Deivedi, S.K.; Hashim, S.R.; Singhal, R. Synthesis and antimicrobial activity of some new quinoxaline derivatives. Pharmaceuticals 2010, 3, 2416–2425. [Google Scholar] [CrossRef]
- Zarranz, B.; Jaso, A.; Aldana, I.; Monge, A. Synthesis and antimycobacterial activity of new quinoxaline-2-carboxamide 1, 4-di-N-oxide derivatives. Bioorg. Med. Chem. 2003, 11, 2149–2156. [Google Scholar] [CrossRef] [Green Version]
- Pereira, J.A.; Pessoa, A.M.; Cordeiro, M.N.D.; Fernandes, R.; Prudencio, C.; Noronha, J.P.; Vieira, M.; Cordeiro, M.N.D.S. Quinoxaline, its derivatives and applications: A State of the Art review. Eur. J. Med. Chem. 2015, 97, 664–672. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, P.; Ganapaty, S.; Rao, C.B. In vitro antitubercular and antimicrobial activities of 1-substituted quinoxaline-2,3(1H,4H)-diones. Bioorg Med. Chem. Lett. 2010, 20, 406–408. [Google Scholar] [CrossRef]
- Petronijević, J.; Janković, N.; Stanojković, T.P.; Joksimović, N.; Grozdanić, N.Đ.; Vraneš, M.; Tot, A.; Bugarčić, Z. Biological evaluation of selected 3,4-dihydro-2(1H)-quinoxalinones and 3,4-dihydro-1,4-benzoxazin-2-ones: Molecular docking study. Arch. Pharm. 2018, 351, 1700308. [Google Scholar] [CrossRef] [PubMed]
- Gein, V.L.; Rassudikhina, N.A.; Shepelina, N.V.; Vakhrin, M.I.; Babushkina, E.B.; Voronina, E.V. Reaction of substituted o-aminophenols with acylpyruvic acid esters and α-ketoglutaric acid. Antibacterial activity of the products. Pharm. Chem. J. 2008, 42, 529–532. [Google Scholar] [CrossRef]
- Kulakov, I.V.; Karbainova, A.A.; Shulgau, Z.T.; Seilkhanov, T.M.; Gatilov, Y.V.; Fisyuk, A.S. Synthesis and Analgesic Activity of bis(3,4-dihydroquinoxalin-2(1H)-one) and bis(3,4-dihydro-2H-1,4-benzoxazin-2-one) Derivatives. Chem. Heterocycl. Compd. 2017, 53, 1094–1097. [Google Scholar] [CrossRef]
- Oleshchuk, A.L.; Shulgau, Z.T.; Seilkhanov, T.M.; Vasilchenko, A.S.; Talipov, S.A.; Kulakov, I.V. Synthesis and Biological Activity of 4-(Pyridin-3-yl)-2-hydroxy-4-oxobut-2-enoic Acid Derivatives. Synlett 2020, 31, 165–170. [Google Scholar] [CrossRef]
- Smith, M.R.; Schirtzinger, E.E.; Wilson, W.C.; Davis, A.S. Rift Valley fever virus: Propagation, quantification, and storage. Curr. Protoc. Microbiol. 2019, 55, e92. [Google Scholar] [CrossRef]
- Puhl, A.C.; Fritch, E.J.; Lane, T.R.; Tse, L.V.; Yount, B.L.; Sacramento, C.Q.; Ekins, S. Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine, and Pyronaridine: In Vitro Activity against SARS-CoV-2 and Potential Mechanisms. ACS Omega 2021, 6, 7454–7468. [Google Scholar] [CrossRef]
- Xiao, X.; Wang, C.; Chang, D.; Wang, Y.; Dong, X.; Jiao, T.; Wang, J. Identification of potent and safe antiviral therapeutic candidates against SARS-CoV-2. Front. Immunol. 2020, 11, 586572. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- BIOVIA. Dassault Systemes, v20. 1. 0. 19295; BIOVIA Discovery Studio, Dassault Systemes: San Diego, CA, USA, 2020. [Google Scholar]
- Available online: https://www.rcsb.org (accessed on 23 May 2022).
- Yan, K.; Rawle, D.J.; Le, T.T.; Suhrbier, A. Simple rapid in vitro screening method for SARS-CoV-2 anti-virals that identifies potential cytomorbidity-associated false positives. Virol. J. 2021, 18, 123. [Google Scholar] [CrossRef]
- Alm, E.; Broberg, E.K.; Connor, T.; Hodcroft, E.B.; Komissarov, A.B.; Maurer-Stroh, S.; Melidou, A.; Neher, R.A.; O’Toole, Á.; Pereyaslov, D. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance 2020, 25, 2001410. [Google Scholar] [CrossRef]
- Case, J.B.; Bailey, A.L.; Kim, A.S.; Chen, R.E.; Diamond, M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 2020, 548, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Brandolini, M.; Taddei, F.; Marino, M.M.; Grumiro, L.; Scalcione, A.; Turba, M.E.; Gentilini, F.; Fantini, M.; Zannoli, S.; Dirani, G.; et al. Correlating qRT-PCR, dPCR and viral titration for the identification and quantification of sars-cov-2: A new approach for infection management. Viruses 2021, 13, 1022. [Google Scholar] [CrossRef] [PubMed]
- Postnikova, E.; Cong, Y.; DeWald, L.E.; Dyall, J.; Yu, S.; Hart, B.J.; Zhou, H.; Gross, R.; Logue, J.; Cai, Y.; et al. Testing therapeutics in cell-based assays: Factors that influence the apparent potency of drugs. PLoS ONE 2018, 13, e0194880. [Google Scholar]
- Motulsky, H.; Christopoulos, A. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting; Oxford University Press: New York, NY, USA, 2004. [Google Scholar]
- Al-Jabri, A.A.; Wigg, M.D.; Oxford, J.S. Initial in vitro screening of drug candidates for their potential antiviral activities. Virol. Methods Man. 1996, 293–308. [Google Scholar] [CrossRef]
- GraphPad Prism 9 User Guide. Available online: http://www.graphpad.com (accessed on 2 January 2021).









| Compound | Structure | IC50, µg/mL * | EC50, µg/mL * |
|---|---|---|---|
| 3 |  | 131.1 | >200 |
| 4a |  | >200 | >200 |
| 4b |  | >200 | >200 |
| 5 |  | >200 | >200 |
| 6a |  | 6.0 | 2.2 |
| 6b |  | >200 | >200 |
| 6c |  | >200 | >200 |
| Tilorone ** |  | 7.8 | 4.3 |
| Receptor | 7AAP | |
|---|---|---|
| Ligand | ||
| 6a | −9.7 | |
| Tilorone | −6.7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stalinskaya, A.L.; Martynenko, N.V.; Shulgau, Z.T.; Shustov, A.V.; Keyer, V.V.; Kulakov, I.V. Synthesis and Antiviral Properties against SARS-CoV-2 of Epoxybenzooxocino[4,3-b]Pyridine Derivatives. Molecules 2022, 27, 3701. https://doi.org/10.3390/molecules27123701
Stalinskaya AL, Martynenko NV, Shulgau ZT, Shustov AV, Keyer VV, Kulakov IV. Synthesis and Antiviral Properties against SARS-CoV-2 of Epoxybenzooxocino[4,3-b]Pyridine Derivatives. Molecules. 2022; 27(12):3701. https://doi.org/10.3390/molecules27123701
Chicago/Turabian StyleStalinskaya, Alena L., Nadezhda V. Martynenko, Zarina T. Shulgau, Alexandr V. Shustov, Viktoriya V. Keyer, and Ivan V. Kulakov. 2022. "Synthesis and Antiviral Properties against SARS-CoV-2 of Epoxybenzooxocino[4,3-b]Pyridine Derivatives" Molecules 27, no. 12: 3701. https://doi.org/10.3390/molecules27123701
APA StyleStalinskaya, A. L., Martynenko, N. V., Shulgau, Z. T., Shustov, A. V., Keyer, V. V., & Kulakov, I. V. (2022). Synthesis and Antiviral Properties against SARS-CoV-2 of Epoxybenzooxocino[4,3-b]Pyridine Derivatives. Molecules, 27(12), 3701. https://doi.org/10.3390/molecules27123701