Gangliosidome of a Human Hippocampus in Temporal Lobe Epilepsy Resolved by High-Resolution Tandem Mass Spectrometry
Abstract
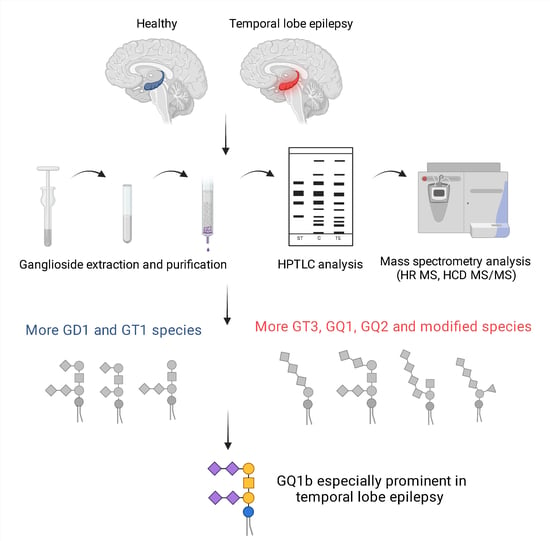
:1. Introduction
2. Results
2.1. High-Performance Thin-Layer Chromatography Detects Minor Changes in Ganglioside Compositional Pattern of Human Hippocampal Tissue in Temporal Epilepsy Compared to Control Tissue
2.2. Comparative Assay by High-Resolution Mass Spectrometry Demonstrates Higher Degree of Gangliosides Species Sialylation, Expression of Fucogangliosides and Modifications of Tri- and Tetrasialogangliosides in Hippocampal Tissue in Temporal Epilepsy
2.3. Structural Analysis of GQ1 Species by High-Energy Collision-Induced Dissociation MS/MS Reveals Specific Association of GQ1b Isomer in Hippocampus with Temporal Epilepsy
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Svennerholm, L. Gangliosides and synaptic transmission. Adv. Exp. Med. Biol. 1980, 125, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Schnaar, R.L. The Biology of Gangliosides. Adv. Carbohydr. Chem. Biochem. 2019, 76, 113–148. [Google Scholar] [CrossRef]
- Xu, Y.; Sun, J.; Yang, L.; Zhao, S.; Liu, X.; Su, Y.; Zhang, J.; Zhao, M. Gangliosides play important roles in the nervous system by regulating ion concentrations. Neurochem. Res. 2022, 47, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Ledeen, R.; Wu, G. Gangliosides of the Nervous System. Methods Mol. Biol. 2018, 1804, 19–55. [Google Scholar] [CrossRef]
- Rahmann, H.; Probst, W.; Muhleisen, M. Gangliosides and synaptic transmission. Jpn. J. Exp. Med. 1982, 52, 275–286. [Google Scholar] [PubMed]
- Tettamanti, G.; Bonali, F.; Marchesini, S.; Zambotti, V. A new procedure for the extraction, purification and fractionation of brain gangliosides. Biochim. Biophys. Acta 1973, 296, 160–170. [Google Scholar] [CrossRef]
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kracun, I.; Rosner, H.; Cosovic, C.; Stavljenic, A. Topographical atlas of the gangliosides of the adult human brain. J. Neurochem. 1984, 43, 979–989. [Google Scholar] [CrossRef]
- Sipione, S.; Monyror, J.; Galleguillos, D.; Steinberg, N.; Kadam, V. Gangliosides in the Brain: Physiology, Pathophysiology and Therapeutic Applications. Front. Neurosci. 2020, 14, 572965. [Google Scholar] [CrossRef]
- Sarbu, M.; Ica, R.; Zamfir, A.D. Gangliosides as Biomarkers of Human Brain Diseases: Trends in Discovery and Characterization by High-Performance Mass Spectrometry. Int. J. Mol. Sci. 2022, 23, 693. [Google Scholar] [CrossRef]
- Huang, F.; Bailey, L.S.; Gao, T.; Jiang, W.; Yu, L.; Bennett, D.A.; Zhao, J.; Basso, K.B.; Guo, Z. Analysis and Comparison of Mouse and Human Brain Gangliosides via Two-Stage Matching of MS/MS Spectra. ACS Omega 2022, 7, 6403–6411. [Google Scholar] [CrossRef] [PubMed]
- Fukami, Y.; Ariga, T.; Yamada, M.; Yuki, N. Brain Gangliosides in Alzheimer’s Disease: Increased Expression of Cholinergic Neuron-Specific Gangliosides. Curr. Alzheimer Res. 2017, 14, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, K. Role of gangliosides in Alzheimer’s disease. Biochim. Biophys. Acta 2007, 1768, 1943–1951. [Google Scholar] [CrossRef] [Green Version]
- Mlinac, K.; Kalanj Bognar, S. Role of gangliosides in brain aging and neurodegeneration. Transl. Neurosci. 2010, 1, 8. [Google Scholar] [CrossRef]
- Sandhoff, R.; Schulze, H.; Sandhoff, K. Ganglioside Metabolism in Health and Disease. Prog. Mol. Biol. Transl. Sci. 2018, 156, 1–62. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.A.; Cross, H.; Proukakis, C.; Priestman, D.A.; Neville, D.C.; Reinkensmeier, G.; Wang, H.; Wiznitzer, M.; Gurtz, K.; Verganelaki, A.; et al. Infantile-onset symptomatic epilepsy syndrome caused by a homozygous loss-of-function mutation of GM3 synthase. Nat. Genet. 2004, 36, 1225–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fragaki, K.; Ait-El-Mkadem, S.; Chaussenot, A.; Gire, C.; Mengual, R.; Bonesso, L.; Beneteau, M.; Ricci, J.E.; Desquiret-Dumas, V.; Procaccio, V.; et al. Refractory epilepsy and mitochondrial dysfunction due to GM3 synthase deficiency. Eur. J. Hum. Genet. 2013, 21, 528–534. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.K.; Holley, J.A.; Macala, L.J.; Spencer, D.D. Ganglioside changes associated with temporal lobe epilepsy in the human hippocampus. Yale J. Biol. Med. 1987, 60, 107–117. [Google Scholar]
- Sarbu, M.; Dehelean, L.; Munteanu, C.; Ica, R.; Petrescu, A.J.; Zamfir, A.D. Human caudate nucleus exhibits a highly complex ganglioside pattern as revealed by high-resolution multistage Orbitrap MS. J. Carbohydr. Chem. 2019, 38, 21. [Google Scholar] [CrossRef]
- Ica, R.; Petrut, A.; Munteanu, C.V.A.; Sarbu, M.; Vukelic, Z.; Petrica, L.; Zamfir, A.D. Orbitrap mass spectrometry for monitoring the ganglioside pattern in human cerebellum development and aging. J. Mass Spectrom. 2020, 55, e4502. [Google Scholar] [CrossRef]
- Ica, R.; Simulescu, A.; Sarbu, M.; Munteanu, C.V.A.; Vukelic, Z.; Zamfir, A.D. High resolution mass spectrometry provides novel insights into the ganglioside pattern of brain cavernous hemangioma. Anal. Biochem. 2020, 609, 113976. [Google Scholar] [CrossRef] [PubMed]
- Dehelean, L.; Sarbu, M.; Petrut, A.; Zamfir, A.D. Trends in Glycolipid Biomarker Discovery in Neurodegenerative Disorders by Mass Spectrometry. Adv. Exp. Med. Biol. 2019, 1140, 703–729. [Google Scholar] [CrossRef] [PubMed]
- Sarbu, M.; Ica, R.; Petrut, A.; Vukelic, Z.; Munteanu, C.V.A.; Petrescu, A.J.; Zamfir, A.D. Gangliosidome of human anencephaly: A high resolution multistage mass spectrometry study. Biochimie 2019, 163, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Fang, H.; Xiao, K.; Liu, Y.; Xue, B.; Tian, Z. Mass measurement accuracy of the Orbitrap in intact proteome analysis. Rapid Commun. Mass Spectrom. 2016, 30, 1391–1397. [Google Scholar] [CrossRef]
- Vukelic, Z.; Zarei, M.; Peter-Katalinic, J.; Zamfir, A.D. Analysis of human hippocampus gangliosides by fully-automated chip-based nanoelectrospray tandem mass spectrometry. J. Chromatogr. A 2006, 1130, 238–245. [Google Scholar] [CrossRef]
- Wu, G.; Lu, Z.H.; Wang, J.; Wang, Y.; Xie, X.; Meyenhofer, M.F.; Ledeen, R.W. Enhanced susceptibility to kainate-induced seizures, neuronal apoptosis, and death in mice lacking gangliotetraose gangliosides: Protection with LIGA 20, a membrane-permeant analog of GM1. J. Neurosci. 2005, 25, 11014–11022. [Google Scholar] [CrossRef] [Green Version]
- Goldman, J.E.; Reynolds, R. A reappraisal of ganglioside GD3 expression in the CNS. Glia 1996, 16, 291–295. [Google Scholar] [CrossRef]
- Simon, B.M.; Malisan, F.; Testi, R.; Nicotera, P.; Leist, M. Disialoganglioside GD3 is released by microglia and induces oligodendrocyte apoptosis. Cell Death Differ. 2002, 9, 758–767. [Google Scholar] [CrossRef] [Green Version]
- Gershen, L.D.; Zanotti-Fregonara, P.; Dustin, I.H.; Liow, J.S.; Hirvonen, J.; Kreisl, W.C.; Jenko, K.J.; Inati, S.K.; Fujita, M.; Morse, C.L.; et al. Neuroinflammation in Temporal Lobe Epilepsy Measured Using Positron Emission Tomographic Imaging of Translocator Protein. JAMA Neurol. 2015, 72, 882–888. [Google Scholar] [CrossRef] [Green Version]
- Kato, K.; Iwamori, M.; Hirabayashi, Y. Increase of GQ1b in the hippocampus of mice following kindled-seizures. Neurosci. Lett. 2008, 441, 286–290. [Google Scholar] [CrossRef]
- Nakamura, Y.; Morimoto, K.; Okamoto, M. Modification of amygdala kindling by intracerebroventricularly administered gangliosides in rats. Exp. Neurol. 1989, 106, 61–69. [Google Scholar] [CrossRef]
- Fujii, S.; Igarashi, K.; Sasaki, H.; Furuse, H.; Ito, K.; Kaneko, K.; Kato, H.; Inokuchi, J.; Waki, H.; Ando, S. Effects of the mono- and tetrasialogangliosides GM1 and GQ1b on ATP-induced long-term potentiation in hippocampal CA1 neurons. Glycobiology 2002, 12, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Ilic, K.; Lin, X.; Malci, A.; Stojanovic, M.; Puljko, B.; Rozman, M.; Vukelic, Z.; Heffer, M.; Montag, D.; Schnaar, R.L.; et al. Plasma Membrane Calcium ATPase-Neuroplastin Complexes Are Selectively Stabilized in GM1-Containing Lipid Rafts. Int. J. Mol. Sci. 2021, 22, 13590. [Google Scholar] [CrossRef] [PubMed]
- Ajith, A.; Mondal, S.; Chattopadhyay, S.; Kumar, A.; Sthanikam, Y.; Chacko, A.G.; Prabhu, K.; Chacko, G.; Vanjare, H.A.; Rajesh, V.; et al. Mass Spectrometry Imaging Deciphers Dysregulated Lipid Metabolism in the Human Hippocampus Affected by Temporal Lobe Epilepsy. ACS Chem. Neurosci. 2021, 12, 4187–4194. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, L.; Fredman, P. A procedure for the quantitative isolation of brain gangliosides. Biochim. Biophys. Acta 1980, 617, 97–109. [Google Scholar] [CrossRef]
- Mlinac, K.; Fabris, D.; Vukelic, Z.; Rozman, M.; Heffer, M.; Bognar, S.K. Structural analysis of brain ganglioside acetylation patterns in mice with altered ganglioside biosynthesis. Carbohydr. Res. 2013, 382, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Schnaar, R.L. Isolation of glycosphingolipids. Methods Enzymol. 1994, 230, 348–370. [Google Scholar] [CrossRef]
- Wells, M.A.; Dittmer, J.C. The Use of Sephadex for the Removal of Nonlipid Contaminants from Lipid Extracts. Biochemistry 1963, 2, 1259–1263. [Google Scholar] [CrossRef]
- Svennerholm, L. Quantitative estimation of sialic acids. II. A colorimetric resorcinol-hydrochloric acid method. Biochim. Biophys. Acta 1957, 24, 604–611. [Google Scholar] [CrossRef]
- Mlinac-Jerkovic, K.; Ilic, K.; Zjalic, M.; Mandic, D.; Debeljak, Z.; Balog, M.; Damjanovic, V.; Macek Hrvat, N.; Habek, N.; Kalanj-Bognar, S.; et al. Who’s in, who’s out? Re-evaluation of lipid raft residents. J. Neurochem. 2021, 158, 657–672. [Google Scholar] [CrossRef]
- Schnaar, R.L.; Needham, L.K. Thin-layer chromatography of glycosphingolipids. Methods Enzymol. 1994, 230, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, L. Ganglioside designation. Adv. Exp. Med. Biol. 1980, 125, 11. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Costello, C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988, 5, 13. [Google Scholar] [CrossRef]
- Costello, C.E.; Juhasz, P.; Perreault, H. New mass spectral approaches to ganglioside structure determinations. Prog. Brain Res. 1994, 101, 45–61. [Google Scholar] [CrossRef] [PubMed]







| No. | m/zexp | m/ztheor | Proposed Structure | Molecular Ion | ppm |
|---|---|---|---|---|---|
| 1 | 603.7844 | 603.783 | GQ1(d18:1/18:0) | [M − 4H+]4− | 2.32 |
| 2 | 608.7863 | 608.783 | GQ1(d18:1/20:0) | [M − 4H+]4− | 5.43 |
| 3 | 610.7934 | 610.7909 | GQ1(d18:1/20:0) and/or GQ1(d20:1/18:0) | [M − 4H+]4− | 4.10 |
| 4 | 613.7989 | 613.8000 | GQ1(d18:0/22:0) | [M − H2O − 4H+]4− | 1.79 |
| 5 | 614.2888 | 614.2855 | GQ1(t18:1/20:1) | [M − 4H+]4− | 5.37 |
| 6 | 617.8014 | 617.799 | GQ1(d18:1/22:0) | [M − 4H+]4− | 3.89 |
| 7 | 620.8065 | 620.8076 | GQ1(d18:0/24:0) | [M − H2O − 4H+]4− | 1.77 |
| 8 | 624.3059 | 624.3024 | GQ1(d18:1/24:1) | [M − 4H+]4− | 5.61 |
| 9 | 637.9702 | 637.967 | Fuc-GT3(t18:1/16:1) | [M − 4H+ +Na+]3− | 5.02 |
| 10 | 674.8742 | 674.8728 | GM2(d18:1/16:2) | [M − 2H+]2− | 2.08 |
| 11 | 699.0083 | 699.0040 | GT1(d18:1/16:0) | [M − 3H+]3− | 6.15 |
| 12 | 708.3524 | 708.3482 | GT1(d18:1/18:0) | [M − 3H+]3− | 5.93 |
| 13 | 713.0035 | 713.008 | GT1(t18:1/18:1) | [M − 3H+]3− | 6.31 |
| 14 | 714.3526 | 714.3512 | GT1(t18:0/18:0) | [M − 3H+]3− | 1.96 |
| 15 | 717.6958 | 717.692 | GT1(d18:1/20:0) and/or GT1(d20:1/18:0) | [M − 3H+]3− | 5.30 |
| 16 | 720.8998 | 720.8964 | GD3(d18:1/16:0) | [M − 2H+]2− | 4.72 |
| 17 | 722.355 | 722.3522 | GT1(t18:1/20:1) | [M − 3H+]3− | 3.88 |
| 18 | 726.3673 | 726.363 | GT1(d18:1/22:1) | [M − 3H+]3− | 5.92 |
| 19 | 727.0393 | 727.036 | GT1(d18:1/22:0) | [M − 3H+]3− | 4.54 |
| 20 | 731.699 | 731.695 | GT1(t18:1/22:1) | [M − 3H+]3− | 5.47 |
| 21 | 734.9159 | 734.9121 | GD3(d18:1/18:0) | [M − 2H+]2− | 5.18 |
| 22 | 733.9079 | 733.9043 | GD3(d18:1/18:0) | [M − 2H+]2− | 4.91 |
| 23 | 735.7112 | 735.7073 | O-Ac-GT1(d18:0/22:0) | [M − H2O − 3H+]3− | 5.31 |
| 24 | 741.0423 | 741.0389 | O-Ac-GT1(d18:1/22:0) | [M − 3H+]3− | 4.59 |
| 25 | 745.3402 | 745.3465 | GQ2(d18:1/17:0) | [M − 3H+]3− | 8.46 |
| 26 | 746.0239 | 746.0184 | GQ2(d18:1/17:1) | [M − 3H+]3− | 7.37 |
| 27 | 771.9344 | 771.9304 | GM1(d18:1/18:0) | [M − 2H+]2− | 5.19 |
| 28 | 775.9554 | 775.9512 | GD3 (d18:1/24:1) | [M − 2H+]2− | 5.42 |
| 29 | 776.9585 | 776.959 | GD3(d18:1/24:0) | [M − 2H+]2− | 0.64 |
| 30 | 789.9623 | 789.9669 | GD3(d18:1/26:1) | [M − 2H+]2− | 5.83 |
| 31 | 796.0399 | 796.036 | GQ1(d18:1/16:0) | [M − 3H+]3− | 4.90 |
| 32 | 804.7126 | 804.708 | GQ1(d18:1/18:1) | [M − 3H+]3− | 5.72 |
| 33 | 805.3842 | 805.3797 | GQ1(d18:1/18:0) | [M − 3H+]3− | 5.59 |
| 34 | 808.7044 | 808.7079 | GQ1(d18:1/19:2) | [M − 3H+]3− | 4.33 |
| 35 | 812.7114 | 812.707 | GQ1(d18:1/18:0) | [M − 4H+ + Na+]3− | 5.42 |
| 36 | 814.728 | 814.724 | GQ1(d18:1/20:0) and/or GQ1(d20:1/18:0) | [M − 3H+]3− | 4.91 |
| 37 | 819.3877 | 819.3832 | O-Ac-GQ1(d18:1/18:0) | [M − 3H+]3− | 5.49 |
| 38 | 820.7048 | 820.7063 | GQ1(d18:0/18:0) | [M − 5H+ + 2Na+]3− | 1.83 |
| 39 | 822.0551 | 822.0508 | GQ1(d18:1/20:0) and/or GQ1(d20:1/18:0) | [M − 4H+ + Na+]3− | 5.23 |
| 40 | 824.0505 | 824.0433 | O-Ac-GQ1(t18:1/18:1) | [M − 3H+]3− | 8.74 |
| 41 | 828.7063 | 828.7028 | (CH3COO−) GQ1(t18:1/18:3) | [M − 3H+]3− | 4.23 |
| 42 | 830.0556 | 830.05 | GQ1(d18:0/20:0) | [M − 5H+ + 2Na+]3− | 6.75 |
| 830.0556 | 830.0509 | GQ1(d18:1/22:2) | [M − 4H+ + Na+]3 | 5.66 | |
| 43 | 831.4135 | 831.4115 | GD2(d18:1/16:2) | [M − 3H+ + Na+]2− | 2.41 |
| 44 | 832.7429 | 832.7391 | GQ1(t18:1/24:0) | [M − H2O − 3H+]3− | 4.57 |
| 45 | 835.4501 | 835.4439 | GD2(d18:1/18:1) | [M − 2H+]2− | 7.43 |
| 46 | 836.456 | 836.452 | GD2(d18:1/18:0) | [M − 2H+]2− | 4.78 |
| 47 | 846.4407 | 846.4349 | GD2(d18:1/18:1) | [M − 3H+ + Na+]2− | 6.86 |
| 48 | 850.4719 | 850.467 | GD2(d18:1/20:0) and/or GD2(d20:1/18:0) | [M − 2H+]2− | 5.76 |
| 49 | 877.4571 | 877.4532 | (CH3COO−) GD2(d18:1/18:0) | [M − 3H+ + Na+]2− | 4.45 |
| 50 | 891.4515 | 891.4508 | GT3(d18:1/18:0) | [M − 3H+ + Na+]2− | 0.79 |
| 51 | 903.4677 | 903.462 | GD1(d18:1/16:0) | [M − 2H+]2− | 6.31 |
| 53 | 916.4758 | 916.4705 | GD1(d18:1/18:1) | [M − 2H+]2− | 5.79 |
| 54 | 917.4836 | 917.478 | GD1(d18:1/18:0) | [M − 2H+]2− | 6.11 |
| 55 | 924.4726 | 924.4678 | GD1(t18:1/18:1) | [M − 2H+]2− | 5.19 |
| 56 | 927.4695 | 927.4613 | GD1(d18:1/18:1) | [M − 3H+ + Na+]2− | 8.85 |
| 57 | 928.4746 | 928.473 | GD1(d18:1/18:0) | [M − 3H+ + Na+]2− | 1.72 |
| 58 | 930.4907 | 930.486 | GD1(d18:1/20:1) | [M − 2H+]2− | 5.05 |
| 59 | 931.4988 | 931.494 | GD1(d18:1/20:0) and/or GD1(d20:1/18:0) | [M − 2H+]2− | 5.16 |
| 60 | 938.4883 | 938.4834 | GD1(t18:1/20:1) | [M − 2H+]2− | 5.22 |
| 61 | 939.4917 | 939.4912 | GD1(t18:1/20:0) | [M − 2H+]2− | 0.53 |
| 62 | 942.4897 | 942.4848 | GD1(d18:1/20:0) and/or GD1(d20:1/18:0) | [M − 3H+ + Na+]2− | 5.20 |
| 63 | 944.5055 | 944.5016 | GD1(d18:1/22:1) | [M − 2H+]2− | 4.13 |
| 64 | 945.5143 | 945.5102 | GD1 (d18:1/22:0) or | [M − 2H+]2− | 4.34 |
| 65 | 945.4976 | O-Ac-GT3(d18:1/24:0) | [M − H2O − 3H+ +Na+]2− | 17.67 | |
| 66 | 946.4793 | 946.4808 | (CH3COO−) GD1(d18:1/18:1) | [M − H+]2− | 1.59 |
| 67 | 951.5007 | 951.4912 | O-Ac-GD1(d18:1/20:1) | [M − 2H+]2− | 9.99 |
| 68 | 954.5082 | 954.5147 | GD1(t18:0/22:0) | [M − 2H+]2− | 6.81 |
| 69 | 957.4667 | 957.4537 | Fuc-GT3(t18:1/16:1) | [M − 3H+ + Na+]2− | 13.58 |
| 70 | 958.4738 | 958.4798 | (CH3COO−) GD1(d18:1/18:0) | [M − 2H+ + Na+]2− | 6.26 |
| 71 | 958.5225 | 958.5173 | GD1(d18:1/24:1) | [M − 2H+]2− | 5.43 |
| 72 | 967.4978 | 967.504 | Fuc-GT3(d18:1/20:0) | [M − 2H+]2− | 6.41 |
| 73 | 968.4676 | 968.4628 | (CH3COO) GD1(d18:1/18:1) | [M − 4H+ + 2Na+]2− | 4.96 |
| 74 | 979.9872 | 979.9865 | GT2(d18:0/16:0) | [M − 3H+ + Na+]2− | 0.72 |
| 75 | 983.4586 | 983.4682 | Fuc-GT3(t18:1/18:0) | [M − 4H+ + 2Na+]2− | 9.77 |
| 76 | 985.4728 | 985.4668 | Fuc-GD1(d18:1/18:2) | [M − 3H+ + Na+]2− | 6.09 |
| 77 | 987.4903 | 987.4837 | Fuc-GD1(d18:1/18:3) | [M − 2H+]2− | 6.69 |
| 78 | 988.4889 | 988.4915 | Fuc-GD1(d18:1/18:2) | [M − 2H+]2− | 2.63 |
| 79 | 989.4889 | 989.4999 | Fuc-GD1(d18:1/18:1) | [M − 2H+]2− | 11.12 |
| 80 | 990.5087 | 990.5071 | Fuc-GD1(d18:1/18:0) | [M − 2H+]2− | 1.62 |
| 81 | 999.519 | 999.5123 | Fuc-GD1(t18:0/18:0) | [M − 2H+]2− | 6.71 |
| 82 | 1007.4932 | 1007.498 | Fuc-(CH3COO−) GT3(d18:1/20:1) | [M − 2H+ + Na+]2− | 4.77 |
| 83 | 1012.4927 | 1012.4903 | Fuc-GD1(d18:1/20:3) | [M − 3H+ + Na+]2− | 2.37 |
| 84 | 1017.9457 | 1017.9555 | GT1(d18:1/12:3) | [M − 2H+]2− | 9.64 |
| 85 | 1019.0033 | 1019.006 | Fuc-(CH3COO−) GD1(d18:1/18:2) | [M − H+]2− | 2.65 |
| 86 | 1033.9827 | 1033.987 | GT1(d18:1/14:1) | [M − 2H+]2− | 4.16 |
| 87 | 1063.0317 | 1063.026 | GT1(d18:1/14:1) | [M − 2H+]2− | 5.36 |
| 88 | 1074.0124 | 1074.017 | GT1(d18:1/18:0) | [M − 3H+ + Na+]2− | 4.28 |
| 89 | 1077.0471 | 1077.042 | GT1(d18:1/20:0) and/or GT1(d20:1/18:0 | [M − 2H+]2− | 4.74 |
| 90 | 1084.0365 | 1084.0311 | GT1(t18:1/20:1) | [M − 2H+]2− | 4.98 |
| 91 | 1087.0289 | 1087.026 | GT1(d18:1/22:4) | [M − 2H+]2− | 2.67 |
| 92 | 1088.0368 | 1088.034 | GT1(d18:1/22:3) | [M − 2H+]2 | 2.57 |
| 93 | 1095.0025 | 1094.9923 | GT1(d18:1/20:3) | [M − 4H+ + 2Na+]2− | 9.32 |
| 94 | 1098.0124 | 1098.0157 | GT1(d18:1/20:1) | [M − 4H+ + 2Na+]2− | 3.01 |
| 95 | 1103.0093 | 1103.02 | (CH3COO−) GT1(d18:1/18:1) | [M − 2H+ + Na+]2− | 9.70 |
| 96 | 1109.0428 | 1109.0365 | GT1(t18:1/20:0) | [M − 4H+ + 2Na+]2− | 5.68 |
| 97 | 1115.0611 | 1115.056 | GT1(d18:1/24:1) | [M − 3H+ + Na+]2− | 4.57 |
| 98 | 1117.077 | 1117.0716 | GT1(d18:0/24:0) | [M − 3H+ + Na+]2− | 4.83 |
| 99 | 1208.5647 | 1208.574 | GQ1(d18:1/18:0) | [M − 2H+]2 | 7.70 |
| 100 | 1218.5552 | 1218.557 | GQ1(d18:1/18:1) | [M − 3H+ + Na+]2− | 1.48 |
| 101 | 1219.5696 | 1219.565 | GQ1(d18:1/18:0) | [M − 3H+ + Na+]2− | 3.77 |
| 102 | 1223.5895 | 1223.597 | GQ1(d18:0/20:0) | [M − 2H+]2 | 6.13 |
| 103 | 1244.577 | 1244.571 | GQ1(d18:1/20:0) and/or GQ1(d20:1/18:0) | [M − 4H+ + 2Na+]2− | 4.82 |
| 104 | 1544.8766 | 1544.869 | GM1(d18:1/18:0) | [M − H+]− | 4.92 |
| 105 | 1572.9064 | 1572.917 | GM1(d18:1/20:0) | [M − H+]− | 6.74 |
| No. | m/zexp | m/ztheor | Proposed Structure | Molecular Ion | ppm |
|---|---|---|---|---|---|
| 1 | 610.7952 | 610.791 | GQ1(d18:1/20:0) | [M − 4H+]4− | 6.89 |
| 2 | 614.2895 | 614.2855 | GQ1(t18:1/20:1) | [M − 4H+]4− | 6.51 |
| 3 | 643.3823 | 643.3802 | GM3(d18:1/24:1) | [M − H2O − 4H+ + 2Na+]3− | 3.27 |
| 4 | 683.6524 | 683.657 | GT1(d18:1/14:0) | [M − 3H+]3− | 6.73 |
| 5 | 699.0085 | 699.004 | GT1(d18:1/16:0) | [M − 3H+]3− | 6.44 |
| 6 | 702.3481 | 702.3444 | GT1(d18:1/18:0) | [M − H2O − 3H+]3− | 5.27 |
| 7 | 708.3528 | 708.3482 | GT1(d18:1/18:0) | [M − 3H+]3− | 6.50 |
| 8 | 714.3559 | 714.3512 | GT1(t18:0/18:0) | [M − 3H+]3− | 6.58 |
| 9 | 717.6961 | 717.692 | GT1(d18:1/20:0) and/or GT1(d20:1/18:0) | [M − 3H+]3− | 5.72 |
| 10 | 720.9006 | 720.8964 | GD3(d18:1/16:0) | [M − 2H+]2− | 5.83 |
| 11 | 722.3559 | 722.3522 | GT1(t18:1/20:1) | [M − 3H+]3− | 5.12 |
| 12 | 727.0396 | 727.036 | GT1(d18:1/22:0) | [M − 3H+]3− | 4.95 |
| 13 | 731.6997 | 731.695 | GT1(t18:1/22:1) | [M − 3H+]3− | 6.43 |
| 14 | 733.9087 | 733.9043 | GD3(d18:1/18:1) | [M − 2H+]2− | 6.00 |
| 15 | 734.9164 | 734.9121 | GD3(d18:1/18:0) | [M − 2H+]2− | 5.86 |
| 16 | 735.6994 | 735.7073 | O-Ac-GT1(d18:0/22:0) | [M − H2O − 3H+]3− | 10.75 |
| 17 | 748.9321 | 748.9277 | GD3(d18:1/20:0) and/or GD3(d20:1/18:0) | [M − 2H+]2− | 5.88 |
| 18 | 789.964 | 789.9669 | GD3(d18:1/26:1) | [M − 2H+]2− | 3.68 |
| 19 | 804.7145 | 804.708 | GQ1(d18:1/18:1) | [M − 3H+]3− | 8.08 |
| 20 | 805.3848 | 805.3797 | GQ1(d18:1/18:0) | [M − 3H+]3− | 6.34 |
| 21 | 812.7127 | 812.707 | GQ1(d18:1/18:0) | [M − 4H+ + Na+]3− | 7.02 |
| 22 | 814.7285 | 814.724 | GQ1(d18:1/20:0) and/or GQ1(d20:1/18:0) | [M − 3H+]3− | 5.53 |
| 23 | 818.7164 | 818.7113 | O-Ac-GQ1(d18:1/18:1) | [M − 3H+]3− | 6.23 |
| 24 | 819.3885 | 819.3832 | O-Ac-GQ1(d18:1/18:0) | [M − 3H+]3− | 6.47 |
| 25 | 822.0567 | 822.0508 | GQ1(d18:1/20:0) and/or GQ1(d20:1/18:0) | [M − 4H+ + Na+]3− | 7.18 |
| 26 | 836.4568 | 836.452 | GD2(d18:1/18:0) | [M − 2H+]2− | 5.74 |
| 27 | 850.4725 | 850.467 | GD2(d18:1/20:0) and/or GD2(d20:1/18:0) | [M − 2H+]2− | 6.47 |
| 28 | 857.472 | 857.4752 | GD2(d18:1/21:0) | [M − 2H+]2 | 3.73 |
| 29 | 865.4962 | 865.4908 | GD2(d18:0/22:0) | [M − 2H+]2 | 6.24 |
| 30 | 880.4648 | 880.4597 | GT3(t18:0/18:0) | [M − H2O − 2H+]2− | 5.80 |
| 31 | 903.4683 | 903.462 | GD1(d18:1/16:0) | [M − 2H+]2− | 6.98 |
| 32 | 916.4763 | 916.4705 | GD1(d18:1/18:1) | [M − 2H+]2− | 6.33 |
| 33 | 917.484 | 917.478 | GD1(d18:1/18:0) | [M − 2H+]2− | 6.54 |
| 34 | 924.4712 | 924.4678 | GD1(t18:1/18:1) | [M − 2H+]2− | 3.68 |
| 35 | 926.489 | 926.4834 | GD1(t18:0/18:0) | [M − 2H+]2− | 6.05 |
| 36 | 930.4915 | 930.486 | GD1(d18:1/20:1) | [M − 2H+]2− | 5.91 |
| 37 | 931.4995 | 931.494 | GD1(d18:1/20:0) and/or GD1(d20:1/18:0) | [M − 2H+]2− | 5.91 |
| 38 | 938.4892 | 938.4834 | GD1(t18:1/20:1) | [M − 2H+]2− | 6.18 |
| 39 | 940.5046 | 940.499 | GD1(t18:0/20:0) | [M − 2H+]2− | 5.96 |
| 40 | 945.5149 | 945.5102 | GD1 (d18:1/22:0) | [M − 2H+]2− | 4.97 |
| 41 | 946.4831 | 946.4808 | (CH3COO−) GD1(d18:1/18:1) | [M − H+]2− | 2.43 |
| 42 | 951.4943 | 951.4912 | O-Ac-GD1(d18:1/20:1) | [M − 2H+]2− | 3.26 |
| 43 | 952.5049 | 952.499 | O-Ac-GD1(d18:1/20:0) | [M − 2H+]2− | 6.20 |
| 44 | 955.4321 | 955.4381 | Fuc-GT3(t18:1/16:3) | [M − 3H+ + Na+]2− | 6.28 |
| 45 | 960.5486 | 960.548 | GD1 (d18:1/24:0) | [M − 2H+]2− | 0.62 |
| 46 | 967.5075 | 967.5044 | Fuc-GT3(d18:1/20:0) | [M − 2H+]2− | 3.21 |
| 47 | 969.4736 | 969.4706 | (CH3COO−) GD1(d18:1/18:0) | [M − 3H+ + 2Na+]2− | 3.10 |
| 48 | 975.4827 | 975.4835 | (CH3COO−)2 GD1(d18:1/18:2) | [M − 2H+]2− | 0.82 |
| 49 | 985.4627 | 985.4668 | Fuc-GD1(d18:1/16:2) | [M − 3H+ + Na+]2− | 4.16 |
| 50 | 987.4895 | 987.4837 | Fuc-GD1(d18:1/18:3) | [M − 2H+]2− | 5.88 |
| 51 | 989.499 | 989.4999 | Fuc-GD1(d18:1/18:1) | [M − 2H+]2− | 0.91 |
| 52 | 990.5132 | 990.5078 | Fuc-GD1(d18:1/18:0) | [M − 2H+]2− | 5.45 |
| 53 | 991.5171 | 991.515 | Fuc-GD1(d18:0/18:0) | [M − 2H+]2− | 2.12 |
| 54 | 992.5197 | 992.512 | Fuc-GT3(d18:1/24:3) | [M − 2H+]2− | 7.76 |
| 55 | 999.5188 | 999.5123 | Fuc-GD1(t18:0/18:0) | [M − 2H+]2− | 6.51 |
| 56 | 1004.522 | 1004.523 | Fuc-GD1(d18:1/20:0) | [M − 2H+]2− | 1.00 |
| 57 | 1007.4842 | 1007.482 | (CH3COO−) GD1(d18:1/22:1) | [M − 4H+ + 3Na+]2− | 2.18 |
| 58 | 1009.5175 | 1009.512 | (CH3COO−) Fuc-GT3(d18:1/22:2) | [M − 2H+]2− | 5.45 |
| 59 | 1013.4927 | 1013.496 | Fuc-GD1(d18:1/20:2) | [M − 3H+ + Na+]2 | 3.26 |
| 60 | 1014.4995 | 1014.506 | Fuc-GD1(d18:1/20:1) | [M − 3H+ + Na+]2 | 6.41 |
| 61 | 1018.9581 | 1018.96 | GT1(d18:1/12:2) | [M − 2H+]2− | 1.87 |
| 62 | 1025.5497 | 1025.544 | Fuc-GD1(d18:1/23:0) | [M − 2H+]2− | 5.56 |
| 63 | 1049.0169 | 1049.0102 | GT1(d18:1/16:0) | [M − 2H+]2− | 6.39 |
| 64 | 1053.9823 | 1053.9842 | GT1(t18:1/16:3) | [M − 2H+]2− | 1.80 |
| 65 | 1060.0079 | 1060.0010 | GT1(d18:1/16:0) | [M − 3H+ + Na+]2− | 6.51 |
| 66 | 1062.025 | 1062.018 | GT1(d18:1/18:1) | [M − 2H+]2− | 6.59 |
| 67 | 1063.0329 | 1063.0260 | GT1(d18:1/18:0) | [M − 2H+]2− | 6.49 |
| 68 | 1074.0231 | 1074.018 | GT1(d18:1/20:3) | [M − 2H+]2− | 4.75 |
| 69 | 1077.0478 | 1077.042 | GT1(d18:1/20:0) and/or GT1(d20:1/18:0) | [M − 2H+]2− | 5.39 |
| 70 | 1081.0081 | 1081.0065 | GT1(t18:1/18:1) | [M − 3H+ + Na+]2− | 1.48 |
| 71 | 1083.0008 | 1082.992 | GT1(d18:1/18:2) | [M − 4H+ + 2Na+]2− | 8.13 |
| 72 | 1084.0375 | 1084.031 | GT1(t18:1/20:1) | [M − 2H+]2− | 6.00 |
| 73 | 1085.0406 | 1085.039 | GT1(t18:1/20:0) | [M − 2H+]2− | 1.47 |
| 74 | 1086.0534 | 1086.047 | GT1(t18:0/20:0) | [M − 2H+]2− | 5.89 |
| 75 | 1088.0382 | 1088.034 | GT1(d18:1/22:3) | [M − 2H+]2− | 3.86 |
| 76 | 1091.0628 | 1091.0579 | GT1(d18:1/22:0) | [M − 2H+]2− | 4.49 |
| 77 | 1095.0281 | 1095.023 | GT1(t18:1/22:4) | [M − 2H+]2− | 4.66 |
| 78 | 1097.0444 | 1097.039 | GT1(t18:1/22:2) | [M − 2H+]2− | 4.92 |
| 79 | 1102.0544 | 1102.048 | GT1(d18:1/22:0) | [M − 3H+ + Na+]2− | 5.81 |
| 80 | 1103.0102 | 1103.02 | (CH3COO−) GT1(d18:1/18:1) | [M − 2H+ + Na+]2− | 8.88 |
| 81 | 1109.0438 | 1109.0390 | GT1(t18:1/24:4) | [M − 2H+]2− | 4.33 |
| 82 | 1112.061 | 1112.0624 | GT1(t18:1/24:1) | [M − 2H+]2− | 1.26 |
| 83 | 1114.0818 | 1114.078 | GT1(t18:0/24:0) | [M − 2H+]2− | 3.41 |
| 84 | 1117.0779 | 1117.0716 | GT1(d18:0/24:0) | [M − 3H+ + 2Na+]2− | 7.43 |
| 85 | 1208.5808 | 1208.574 | GQ1(d18:1/18:0) | [M − 2H+]2− | 5.63 |
| 86 | 1218.5662 | 1218.557 | GQ1(d18:1/18:1) | [M − 3H+ + Na+]2− | 7.55 |
| 87 | 1219.5713 | 1219.565 | GQ1(d18:1/18:0) | [M − 3H+ + Na+]2− | 5.17 |
| 88 | 1222.5967 | 1222.5899 | GQ1(d18:1/20:0) | [M − 2H+]2− | 5.56 |
| 89 | 1229.5871 | 1229.579 | O-Ac-GQ1(d18:1/18:0) | [M − 2H+]2− | 6.59 |
| 90 | 1244.5784 | 1244.571 | GQ1(d18:1/20:0) and/or GQ1(d20:1/18:0) | [M − 4H+ + 2Na+]2− | 5.95 |
| 91 | 1544.8765 | 1544.869 | GM1(d18:1/18:0) | [M − H+]− | 4.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ica, R.; Mlinac-Jerkovic, K.; Ilic, K.; Sajko, T.; Munteanu, C.V.A.; Zamfir, A.D.; Kalanj-Bognar, S. Gangliosidome of a Human Hippocampus in Temporal Lobe Epilepsy Resolved by High-Resolution Tandem Mass Spectrometry. Molecules 2022, 27, 4056. https://doi.org/10.3390/molecules27134056
Ica R, Mlinac-Jerkovic K, Ilic K, Sajko T, Munteanu CVA, Zamfir AD, Kalanj-Bognar S. Gangliosidome of a Human Hippocampus in Temporal Lobe Epilepsy Resolved by High-Resolution Tandem Mass Spectrometry. Molecules. 2022; 27(13):4056. https://doi.org/10.3390/molecules27134056
Chicago/Turabian StyleIca, Raluca, Kristina Mlinac-Jerkovic, Katarina Ilic, Tomislav Sajko, Cristian V. A. Munteanu, Alina D. Zamfir, and Svjetlana Kalanj-Bognar. 2022. "Gangliosidome of a Human Hippocampus in Temporal Lobe Epilepsy Resolved by High-Resolution Tandem Mass Spectrometry" Molecules 27, no. 13: 4056. https://doi.org/10.3390/molecules27134056
APA StyleIca, R., Mlinac-Jerkovic, K., Ilic, K., Sajko, T., Munteanu, C. V. A., Zamfir, A. D., & Kalanj-Bognar, S. (2022). Gangliosidome of a Human Hippocampus in Temporal Lobe Epilepsy Resolved by High-Resolution Tandem Mass Spectrometry. Molecules, 27(13), 4056. https://doi.org/10.3390/molecules27134056